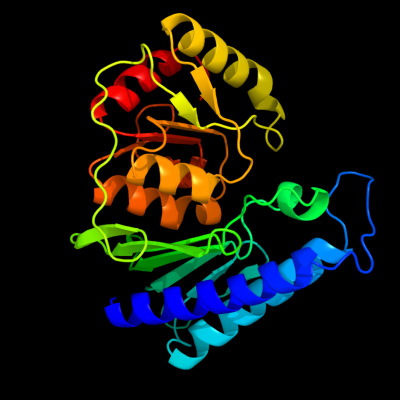
1 c2qflA_
100.0
100
PDB header: hydrolaseChain: A: PDB Molecule: inositol-1-monophosphatase;PDBTitle: structure of suhb: inositol monophosphatase and extragenic2 suppressor from e. coli
2 c2p3nB_
100.0
36
PDB header: hydrolaseChain: B: PDB Molecule: inositol-1-monophosphatase;PDBTitle: thermotoga maritima impase tm1415
3 c3luzA_
100.0
44
PDB header: hydrolaseChain: A: PDB Molecule: extragenic suppressor protein suhb;PDBTitle: crystal structure of extragenic suppressor protein suhb from2 bartonella henselae, via combined iodide sad molecular replacement
4 d2hhma_
100.0
32
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like5 c2czhB_
100.0
33
PDB header: hydrolaseChain: B: PDB Molecule: inositol monophosphatase 2;PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
6 c2fvzB_
100.0
33
PDB header: hydrolaseChain: B: PDB Molecule: inositol monophosphatase 2;PDBTitle: human inositol monophosphosphatase 2
7 c2pcrA_
100.0
35
PDB header: hydrolaseChain: A: PDB Molecule: inositol-1-monophosphatase;PDBTitle: crystal structure of myo-inositol-1(or 4)-monophosphatase (aq_1983)2 from aquifex aeolicus vf5
8 d1g0ha_
100.0
25
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like9 c2q74B_
100.0
35
PDB header: hydrolaseChain: B: PDB Molecule: inositol-1-monophosphatase;PDBTitle: mycobacterium tuberculosis suhb
10 d1jp4a_
100.0
19
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like11 d1ka1a_
100.0
28
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like12 d1vdwa_
100.0
31
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like13 d1xi6a_
100.0
29
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like14 d1lbva_
100.0
31
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like15 c3b8bA_
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: cysq, sulfite synthesis pathway protein;PDBTitle: crystal structure of cysq from bacteroides thetaiotaomicron, a2 bacterial member of the inositol monophosphatase family
16 d1inpa_
100.0
24
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like17 c3uksB_
99.9
18
PDB header: hydrolaseChain: B: PDB Molecule: sedoheptulose-1,7 bisphosphatase, putative;PDBTitle: 1.85 angstrom crystal structure of putative sedoheptulose-1,72 bisphosphatase from toxoplasma gondii
18 d1d9qa_
99.9
19
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like19 d1nuwa_
99.6
16
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like20 c2fhyL_
99.6
12
PDB header: hydrolaseChain: L: PDB Molecule: fructose-1,6-bisphosphatase 1;PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
21 d1ftaa_
not modelled
99.5
15
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like22 d1bk4a_
not modelled
99.4
15
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like23 c2gq1A_
not modelled
99.3
18
PDB header: hydrolaseChain: A: PDB Molecule: fructose-1,6-bisphosphatase;PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
24 d1spia_
not modelled
99.1
19
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like25 d1ni9a_
not modelled
93.5
22
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: GlpX-like bacterial fructose-1,6-bisphosphatase26 d1o12a1
not modelled
58.1
36
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA27 c3fhkF_
not modelled
49.2
8
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: upf0403 protein yphp;PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide2 isomerase
28 d1mdah_
not modelled
32.2
20
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain29 d1r89a1
not modelled
17.0
33
Fold: PAP/OAS1 substrate-binding domainSuperfamily: PAP/OAS1 substrate-binding domainFamily: Archaeal tRNA CCA-adding enzyme substrate-binding domain30 c3mx7A_
not modelled
16.2
20
PDB header: apoptosisChain: A: PDB Molecule: fas apoptotic inhibitory molecule 1;PDBTitle: crystal structure analysis of human faim-ntd
31 c1ikqA_
not modelled
14.3
40
PDB header: transferaseChain: A: PDB Molecule: exotoxin a;PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
32 d2ns0a1
not modelled
14.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like33 d3pmga4
not modelled
12.6
13
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain34 c2o7jA_
not modelled
11.5
21
PDB header: sugar binding proteinChain: A: PDB Molecule: oligopeptide abc transporter, periplasmicPDBTitle: the x-ray crystal structure of a thermophilic cellobiose2 binding protein bound with cellopentaose
35 d2dsqg1
not modelled
11.2
20
Fold: Thyroglobulin type-1 domainSuperfamily: Thyroglobulin type-1 domainFamily: Thyroglobulin type-1 domain36 c2pncB_
not modelled
9.8
23
PDB header: oxidoreductaseChain: B: PDB Molecule: copper amine oxidase, liver isozyme;PDBTitle: crystal structure of bovine plasma copper-containing amine oxidase in2 complex with clonidine
37 d2j9ga2
not modelled
9.7
23
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like38 c3g5uB_
not modelled
9.3
20
PDB header: membrane proteinChain: B: PDB Molecule: multidrug resistance protein 1a;PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
39 d1icfi_
not modelled
8.9
23
Fold: Thyroglobulin type-1 domainSuperfamily: Thyroglobulin type-1 domainFamily: Thyroglobulin type-1 domain40 d2g5gx1
not modelled
8.7
18
Fold: EreA/ChaN-likeSuperfamily: EreA/ChaN-likeFamily: ChaN-like41 d1w96a2
not modelled
7.9
19
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like42 c3b5xB_
not modelled
7.5
16
PDB header: membrane proteinChain: B: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: crystal structure of msba from vibrio cholerae
43 c3equB_
not modelled
7.5
20
PDB header: biosynthetic proteinChain: B: PDB Molecule: penicillin-binding protein 2;PDBTitle: crystal structure of penicillin-binding protein 2 from neisseria2 gonorrhoeae
44 d1kfia4
not modelled
7.4
38
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain45 c2hydB_
not modelled
7.3
18
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
46 c1sz1A_
not modelled
6.9
27
PDB header: transferase/rnaChain: A: PDB Molecule: trna nucleotidyltransferase;PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
47 d1l0wa2
not modelled
6.8
32
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain48 d1g4ma1
not modelled
6.5
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Arrestin/Vps26-like49 c2h7tA_
not modelled
6.1
19
PDB header: protein bindingChain: A: PDB Molecule: insulin-like growth factor-binding protein 2;PDBTitle: solution structure of the c-terminal domain of insulin-like2 growth factor binding protein 2 (igfbp-2)