1 c3ia0c_
100.0
99
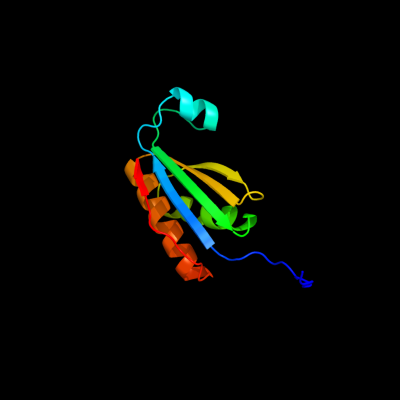
PDB header: structural proteinChain: C: PDB Molecule: ethanolamine utilization protein euts;PDBTitle: ethanolamine utilization microcompartment shell subunit,2 euts-g39v mutant
2 c3cgiD_
100.0
56
PDB header: unknown functionChain: D: PDB Molecule: propanediol utilization protein pduu;PDBTitle: crystal structure of the pduu shell protein from the pdu2 microcompartment
3 c3i82A_
99.8
25
PDB header: structural proteinChain: A: PDB Molecule: ethanolamine utilization protein eutl;PDBTitle: ethanolamine utilization microcompartment shell subunit, eutl closed2 form
4 c3nwgA_
99.4
20
PDB header: structural proteinChain: A: PDB Molecule: microcompartments protein;PDBTitle: the crystal structure of a microcomparments protein from2 desulfitobacterium hafniense dcb
5 c3io0A_
99.4
31
PDB header: structural proteinChain: A: PDB Molecule: etub protein;PDBTitle: crystal structure of etub from clostridium kluyveri
6 c3n79A_
99.3
25
PDB header: electron transportChain: A: PDB Molecule: pdut;PDBTitle: pdut c38s mutant from salmonella enterica typhimurium
7 d2ewha1
98.7
27
Fold: Ferredoxin-likeSuperfamily: CcmK-likeFamily: CcmK-like8 d2a10a1
98.6
23
Fold: Ferredoxin-likeSuperfamily: CcmK-likeFamily: CcmK-like9 c3ngkA_
98.6
24
PDB header: unknown functionChain: A: PDB Molecule: propanediol utilization protein pdua;PDBTitle: pdua from salmonella enterica typhimurium
10 d2a1ba1
98.6
22
Fold: Ferredoxin-likeSuperfamily: CcmK-likeFamily: CcmK-like11 c3i6pF_
98.1
24
PDB header: structural proteinChain: F: PDB Molecule: ethanolamine utilization protein eutm;PDBTitle: ethanolamine utilization microcompartment shell subunit, eutm
12 c3f56F_
93.7
23
PDB header: structural proteinChain: F: PDB Molecule: csos1d;PDBTitle: the structure of a previously undetected carboxysome shell2 protein: csos1d from prochlorococcus marinus med4
13 d2r48a1
80.5
29
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like14 d2r4qa1
79.6
32
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like15 c2kyrA_
77.6
21
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
16 d1qnaa1
69.0
23
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain17 d1nh2a1
66.5
19
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain18 d1cdwa1
61.7
23
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain19 d1qlma_
52.0
20
Fold: Methenyltetrahydromethanopterin cyclohydrolaseSuperfamily: Methenyltetrahydromethanopterin cyclohydrolaseFamily: Methenyltetrahydromethanopterin cyclohydrolase20 c3eikB_
41.2
26
PDB header: transcriptionChain: B: PDB Molecule: tata-box-binding protein;PDBTitle: double stranded dna binding protein
21 d1t1ea2
not modelled
38.2
18
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors22 c1ngmM_
not modelled
36.2
19
PDB header: transcription/dnaChain: M: PDB Molecule: transcription initiation factor tfiid;PDBTitle: crystal structure of a yeast brf1-tbp-dna ternary complex
23 c1rm1A_
not modelled
36.2
19
PDB header: transcription/dnaChain: A: PDB Molecule: tata-box binding protein;PDBTitle: structure of a yeast tfiia/tbp/tata-box dna complex
24 d1nh2a2
not modelled
35.3
23
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain25 c2z8uQ_
not modelled
32.6
30
PDB header: transcriptionChain: Q: PDB Molecule: tata-box-binding protein;PDBTitle: methanococcus jannaschii tbp
26 d1x4ma1
not modelled
28.8
21
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)27 d1mp9a2
not modelled
28.3
33
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain28 c1njjC_
not modelled
26.0
10
PDB header: lyaseChain: C: PDB Molecule: ornithine decarboxylase;PDBTitle: crystal structure determination of t. brucei ornithine2 decarboxylase bound to d-ornithine and to g418
29 c2hh3A_
not modelled
24.5
25
PDB header: rna binding proteinChain: A: PDB Molecule: kh-type splicing regulatory protein;PDBTitle: solution structure of the third kh domain of ksrp
30 d1aisa2
not modelled
23.8
31
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain31 d1cdwa2
not modelled
23.2
27
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain32 c2hh2A_
not modelled
23.0
19
PDB header: rna binding proteinChain: A: PDB Molecule: kh-type splicing regulatory protein;PDBTitle: solution structure of the fourth kh domain of ksrp
33 d1qnaa2
not modelled
21.6
27
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain34 d1we8a_
not modelled
21.3
21
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)35 d1mp9a1
not modelled
19.9
27
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain36 c1t1eA_
not modelled
18.0
18
PDB header: hydrolaseChain: A: PDB Molecule: kumamolisin;PDBTitle: high resolution crystal structure of the intact pro-2 kumamolisin, a sedolisin type proteinase (previously3 called kumamolysin or kscp)
37 c1ztgD_
not modelled
17.5
16
PDB header: dna, rna binding protein/dnaChain: D: PDB Molecule: poly(rc)-binding protein 1;PDBTitle: human alpha polyc binding protein kh1
38 c2jzxA_
not modelled
16.8
17
PDB header: rna binding proteinChain: A: PDB Molecule: poly(rc)-binding protein 2;PDBTitle: pcbp2 kh1-kh2 domains
39 c1mp9B_
not modelled
16.8
31
PDB header: dna binding proteinChain: B: PDB Molecule: tata-binding protein;PDBTitle: tbp from a mesothermophilic archaeon, sulfolobus2 acidocaldarius
40 d1j4wa1
not modelled
16.3
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)41 c3n89B_
not modelled
16.2
12
PDB header: cell cycleChain: B: PDB Molecule: defective in germ line development protein 3, isoform a;PDBTitle: kh domains
42 d1dtja_
not modelled
15.8
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)43 d1khma_
not modelled
14.1
14
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)44 d1s99a_
not modelled
13.6
13
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: Putative thiamin/HMP-binding protein YkoF45 c1mzjB_
not modelled
12.6
18
PDB header: transferaseChain: B: PDB Molecule: beta-ketoacylsynthase iii;PDBTitle: crystal structure of the priming beta-ketosynthase from the2 r1128 polyketide biosynthetic pathway
46 d2axya1
not modelled
12.4
13
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)47 c3ipfA_
not modelled
12.1
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q251q8_deshy protein from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr8c.
48 d1ec6a_
not modelled
11.8
16
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)49 d1zzka1
not modelled
11.4
14
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)50 d1d7ka2
not modelled
11.4
16
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain51 d2ba0a3
not modelled
11.3
19
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)52 c3edyA_
not modelled
11.2
18
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure of the precursor form of human tripeptidyl-peptidase2 1
53 c1iruF_
not modelled
10.7
20
PDB header: hydrolaseChain: F: PDB Molecule: 20s proteasome;PDBTitle: crystal structure of the mammalian 20s proteasome at 2.75 a2 resolution
54 d1iruf_
not modelled
10.7
20
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits55 d1irug_
not modelled
10.5
33
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits56 d2ctma1
not modelled
10.1
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)57 c3bdmF_
not modelled
9.7
27
PDB header: hydrolaseChain: F: PDB Molecule: proteasome component c1;PDBTitle: yeast 20s proteasome:glidobactin a-complex
58 d1rypg_
not modelled
9.7
27
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits59 c3h76A_
not modelled
9.7
18
PDB header: transferaseChain: A: PDB Molecule: pqs biosynthetic enzyme;PDBTitle: crystal structure of pqsd, a key enzyme in pseudomonas2 aeruginosa quinolone signal biosynthesis pathway
60 d1irua_
not modelled
9.4
31
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits61 c1d3uA_
not modelled
9.1
31
PDB header: gene regulation/dnaChain: A: PDB Molecule: tata-binding protein;PDBTitle: tata-binding protein/transcription factor (ii)b/bre+tata-2 box complex from pyrococcus woesei
62 d2ooia1
not modelled
8.7
6
Fold: Chorismate lyase-likeSuperfamily: Chorismate lyase-likeFamily: UTRA domain63 d1rype_
not modelled
8.7
31
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits64 d1x4na1
not modelled
8.6
13
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)65 d2ctka1
not modelled
8.4
18
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)66 d1aisa1
not modelled
8.3
38
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain67 c2qnxA_
not modelled
8.0
16
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: crystal structure of the complex between the mycobacterium beta-2 ketoacyl-acyl carrier protein synthase iii (fabh) and 11-3 [(decyloxycarbonyl)dithio]-undecanoic acid
68 c3df7A_
not modelled
8.0
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative atp-grasp superfamily protein;PDBTitle: crystal structure of a putative atp-grasp superfamily2 protein from archaeoglobus fulgidus
69 d1rypf_
not modelled
7.7
33
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits70 d1rypb_
not modelled
7.7
27
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits71 c2e3uA_
not modelled
7.5
11
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein ph1566;PDBTitle: crystal structure analysis of dim2p from pyrococcus horikoshii ot3
72 d1irud_
not modelled
7.4
27
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits73 c2kp6A_
not modelled
6.8
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein cv0237 from2 chromobacterium violaceum. northeast structural genomics3 consortium (nesg) target cvt1
74 d1iruc_
not modelled
6.8
31
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits75 d1rypa_
not modelled
6.6
38
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits76 d1irub_
not modelled
6.3
27
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits77 d1nvmb2
not modelled
6.0
39
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like78 c3lpeF_
not modelled
6.0
35
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
79 d1rypd_
not modelled
5.8
38
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits80 d1irue_
not modelled
5.8
23
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits81 d2ctla1
not modelled
5.8
33
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)82 c3krmB_
not modelled
5.7
13
PDB header: rna binding proteinChain: B: PDB Molecule: insulin-like growth factor 2 mrna-binding proteinPDBTitle: imp1 kh34
83 c3orsD_
not modelled
5.7
11
PDB header: isomerase,biosynthetic proteinChain: D: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
84 d2ctfa1
not modelled
5.5
21
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)85 d1rypc_
not modelled
5.5
38
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits86 c2pn1A_
not modelled
5.5
21
PDB header: ligaseChain: A: PDB Molecule: carbamoylphosphate synthase large subunit;PDBTitle: crystal structure of carbamoylphosphate synthase large subunit (split2 gene in mj) (zp_00538348.1) from exiguobacterium sp. 255-15 at 2.00 a3 resolution
87 d1wvna1
not modelled
5.4
23
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)88 c2dgrA_
not modelled
5.3
14
PDB header: rna binding proteinChain: A: PDB Molecule: ring finger and kh domain-containing protein 1;PDBTitle: solution structure of the second kh domain in ring finger2 and kh domain containing protein 1
89 c2anrA_
not modelled
5.3
9
PDB header: rna-binding protein/rnaChain: A: PDB Molecule: neuro-oncological ventral antigen 1;PDBTitle: crystal structure (ii) of nova-1 kh1/kh2 domain tandem with 25nt rna2 hairpin