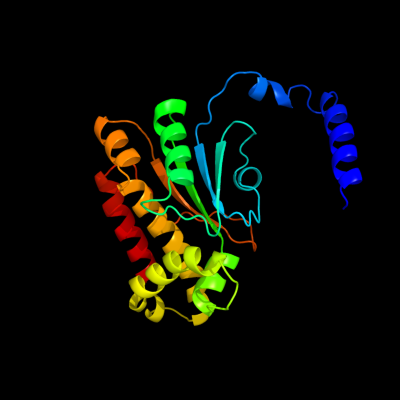
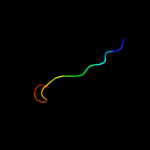
1 d1i6pa_
100.0
100
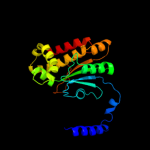
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab2 c2a8cE_
100.0
63
PDB header: lyaseChain: E: PDB Molecule: carbonic anhydrase 2;PDBTitle: haemophilus influenzae beta-carbonic anhydrase
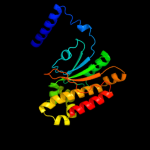
3 d1ddza1
100.0
48
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab4 d1ddza2
100.0
45
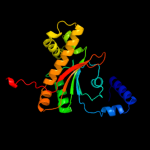
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab5 c1ddzA_
100.0
47
PDB header: lyaseChain: A: PDB Molecule: carbonic anhydrase;PDBTitle: x-ray structure of a beta-carbonic anhydrase from the red2 alga, porphyridium purpureum r-1
6 c3ucoB_
100.0
39
PDB header: lyase/lyase inhibitorChain: B: PDB Molecule: carbonic anhydrase;PDBTitle: coccomyxa beta-carbonic anhydrase in complex with iodide
7 c2w3nA_
100.0
33
PDB header: lyaseChain: A: PDB Molecule: carbonic anhydrase 2;PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
8 d1ekja_
100.0
25
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab9 c3eyxB_
100.0
30
PDB header: lyaseChain: B: PDB Molecule: carbonic anhydrase;PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
10 c2a5vB_
100.0
28
PDB header: lyaseChain: B: PDB Molecule: carbonic anhydrase (carbonate dehydratase) (carbonicPDBTitle: crystal structure of m. tuberculosis beta carbonic anhydrase, rv3588c,2 tetrameric form
11 c3lasA_
100.0
23
PDB header: lyaseChain: A: PDB Molecule: putative carbonic anhydrase;PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
12 c1ylkA_
100.0
18
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein rv1284/mt1322;PDBTitle: crystal structure of rv1284 from mycobacterium tuberculosis in complex2 with thiocyanate
13 d1g5ca_
100.0
24
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab14 c2hwkA_
62.6
26
PDB header: hydrolaseChain: A: PDB Molecule: helicase nsp2;PDBTitle: crystal structure of venezuelan equine encephalitis2 alphavirus nsp2 protease domain
15 d1j2ra_
36.5
19
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases16 c3degC_
30.9
21
PDB header: ribosomeChain: C: PDB Molecule: gtp-binding protein lepa;PDBTitle: complex of elongating escherichia coli 70s ribosome and ef4(lepa)-2 gmppnp
17 c2h5eB_
28.9
21
PDB header: translationChain: B: PDB Molecule: peptide chain release factor rf-3;PDBTitle: crystal structure of e.coli polypeptide release factor rf3
18 c3i8sC_
28.7
27
PDB header: transport proteinChain: C: PDB Molecule: ferrous iron transport protein b;PDBTitle: structure of the cytosolic domain of e. coli feob, nucleotide-free2 form
19 d1n0ua2
28.6
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins20 d1fqta_
28.2
14
Fold: ISP domainSuperfamily: ISP domainFamily: Rieske iron-sulfur protein (ISP)21 c2de7E_
not modelled
28.0
31
PDB header: oxidoreductaseChain: E: PDB Molecule: ferredoxin component of carbazole;PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
22 d1zo0a1
not modelled
27.4
21
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Ornithine decarboxylase antizyme-like23 c3d89A_
not modelled
27.3
17
PDB header: electron transportChain: A: PDB Molecule: rieske domain-containing protein;PDBTitle: crystal structure of a soluble rieske ferredoxin from mus musculus
24 d1nrjb_
not modelled
26.8
42
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins25 d1z01a1
not modelled
26.0
29
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain26 d2de6a1
not modelled
25.7
50
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain27 c2ywfA_
not modelled
25.2
29
PDB header: translationChain: A: PDB Molecule: gtp-binding protein lepa;PDBTitle: crystal structure of gmppnp-bound lepa from aquifex aeolicus
28 d2fh5b1
not modelled
24.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins29 c3trkA_
not modelled
24.5
16
PDB header: hydrolaseChain: A: PDB Molecule: nonstructural polyprotein;PDBTitle: structure of the chikungunya virus nsp2 protease
30 c3ibyA_
not modelled
24.5
33
PDB header: transport proteinChain: A: PDB Molecule: ferrous iron transport protein b;PDBTitle: structure of cytosolic domain of l. pneumophila feob
31 c3gceA_
not modelled
24.3
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin component of carbazole 1,9a-PDBTitle: ferredoxin of carbazole 1,9a-dioxygenase from nocardioides2 aromaticivorans ic177
32 d1rkba_
not modelled
24.2
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases33 c1d2eA_
not modelled
24.2
27
PDB header: rna binding proteinChain: A: PDB Molecule: elongation factor tu (ef-tu);PDBTitle: crystal structure of mitochondrial ef-tu in complex with gdp
34 c2qu8A_
not modelled
23.9
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative nucleolar gtp-binding protein 1;PDBTitle: crystal structure of putative nucleolar gtp-binding protein 1 pff0625w2 from plasmodium falciparum
35 d2jo6a1
not modelled
23.2
25
Fold: ISP domainSuperfamily: ISP domainFamily: NirD-like36 c3p27A_
not modelled
22.9
27
PDB header: signaling proteinChain: A: PDB Molecule: elongation factor 1 alpha-like protein;PDBTitle: crystal structure of a translational gtpase (gdp form)
37 c3k53B_
not modelled
22.5
15
PDB header: metal transportChain: B: PDB Molecule: ferrous iron transport protein b;PDBTitle: crystal structure of nfeob from p. furiosus
38 c1mj1A_
not modelled
22.4
27
PDB header: ribosomeChain: A: PDB Molecule: elongation factor tu;PDBTitle: fitting the ternary complex of ef-tu/trna/gtp and ribosomal proteins2 into a 13 a cryo-em map of the coli 70s ribosome
39 d1s0ua3
not modelled
22.3
36
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins40 c3izyP_
not modelled
21.8
29
PDB header: rna, ribosomal proteinChain: P: PDB Molecule: translation initiation factor if-2, mitochondrial;PDBTitle: mammalian mitochondrial translation initiation factor 2
41 c3tr5C_
not modelled
21.3
21
PDB header: translationChain: C: PDB Molecule: peptide chain release factor 3;PDBTitle: structure of a peptide chain release factor 3 (prfc) from coxiella2 burnetii
42 c2bvnB_
not modelled
21.1
27
PDB header: elongation factorChain: B: PDB Molecule: elongation factor tu;PDBTitle: e. coli ef-tu:gdpnp in complex with the antibiotic2 enacyloxin iia
43 d1r5ba3
not modelled
21.1
36
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins44 c2dy1A_
not modelled
21.0
29
PDB header: signaling protein, translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of ef-g-2 from thermus thermophilus
45 c1zunB_
not modelled
20.7
27
PDB header: transferaseChain: B: PDB Molecule: sulfate adenylate transferase, subunitPDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
46 d1efca3
not modelled
20.1
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins47 c2qagC_
not modelled
20.0
27
PDB header: cell cycle, structural proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of human septin trimer 2/6/7
48 c3oqpB_
not modelled
19.9
11
PDB header: hydrolaseChain: B: PDB Molecule: putative isochorismatase;PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
49 d1g7sa4
not modelled
19.9
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins50 c2rauA_
not modelled
19.3
18
PDB header: hydrolaseChain: A: PDB Molecule: putative esterase;PDBTitle: crystal structure of a putative lipase (np_343859.1) from sulfolobus2 solfataricus at 1.85 a resolution
51 c1r5nA_
not modelled
19.3
36
PDB header: translationChain: A: PDB Molecule: eukaryotic peptide chain release factor gtp-PDBTitle: crystal structure analysis of sup35 complexed with gdp
52 c2zxrA_
not modelled
19.1
24
PDB header: hydrolaseChain: A: PDB Molecule: single-stranded dna specific exonuclease recj;PDBTitle: crystal structure of recj in complex with mg2+ from thermus2 thermophilus hb8
53 c3hb7G_
not modelled
19.1
14
PDB header: hydrolaseChain: G: PDB Molecule: isochorismatase hydrolase;PDBTitle: the crystal structure of an isochorismatase-like hydrolase from2 alkaliphilus metalliredigens to 2.3a
54 d1ulra_
not modelled
19.0
14
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like55 c3t5dC_
not modelled
18.9
27
PDB header: signaling proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of septin 7 in complex with gdp
56 c2qpzA_
not modelled
18.8
15
PDB header: metal binding proteinChain: A: PDB Molecule: naphthalene 1,2-dioxygenase system ferredoxinPDBTitle: naphthalene 1,2-dioxygenase rieske ferredoxin
57 d1svia_
not modelled
18.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins58 c2xexA_
not modelled
18.4
29
PDB header: translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
59 d2ih2a2
not modelled
18.2
24
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: TaqI C-terminal domain-like60 c3br8A_
not modelled
17.6
14
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
61 c3pqcA_
not modelled
17.6
14
PDB header: hydrolaseChain: A: PDB Molecule: probable gtp-binding protein engb;PDBTitle: crystal structure of thermotoga maritima ribosome biogenesis gtp-2 binding protein engb (ysxc/yiha) in complex with gdp
62 c2i7fB_
not modelled
17.5
36
PDB header: oxidoreductaseChain: B: PDB Molecule: ferredoxin component of dioxygenase;PDBTitle: sphingomonas yanoikuyae b1 ferredoxin
63 c1s0uA_
not modelled
17.3
36
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: eif2gamma apo
64 d3c0da1
not modelled
17.1
9
Fold: ISP domainSuperfamily: ISP domainFamily: NirD-like65 c3a1vB_
not modelled
17.0
18
PDB header: transport proteinChain: B: PDB Molecule: iron(ii) transport protein b;PDBTitle: crystal structue of the cytosolic domain of t. maritima feob2 iron iransporter in apo form
66 c1z01D_
not modelled
17.0
27
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-oxo-1,2-dihydroquinoline 8-monooxygenase,PDBTitle: 2-oxoquinoline 8-monooxygenase component: active site2 modulation by rieske-[2fe-2s] center oxidation/reduction
67 d1egaa1
not modelled
16.9
40
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins68 d1vm9a_
not modelled
16.8
27
Fold: ISP domainSuperfamily: ISP domainFamily: Rieske iron-sulfur protein (ISP)69 c2wjjB_
not modelled
16.5
27
PDB header: metal transportChain: B: PDB Molecule: ferrous iron transport protein b homolog;PDBTitle: structure and function of the feob g-domain from2 methanococcus jannaschii
70 d2bv3a2
not modelled
16.5
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins71 c1zo1I_
not modelled
16.3
29
PDB header: translation/rnaChain: I: PDB Molecule: translation initiation factor 2;PDBTitle: if2, if1, and trna fitted to cryo-em data of e. coli 70s2 initiation complex
72 d2jzaa1
not modelled
16.2
18
Fold: ISP domainSuperfamily: ISP domainFamily: NirD-like73 c1g7cA_
not modelled
16.1
27
PDB header: translationChain: A: PDB Molecule: elongation factor 1-alpha;PDBTitle: yeast eef1a:eef1ba in complex with gdpnp
74 c3oqpA_
not modelled
16.0
11
PDB header: hydrolaseChain: A: PDB Molecule: putative isochorismatase;PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
75 d1udxa2
not modelled
15.9
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins76 c2plfA_
not modelled
15.6
36
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: the structure of aif2gamma subunit from the archaeon2 sulfolobus solfataricus in the nucleotide-free form.
77 c2x2fD_
not modelled
15.6
27
PDB header: hydrolaseChain: D: PDB Molecule: dynamin-1;PDBTitle: dynamin 1 gtpase dimer, short axis form
78 c2zovA_
not modelled
15.4
33
PDB header: membrane proteinChain: A: PDB Molecule: chemotaxis protein motb;PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form i)
79 d1apsa_
not modelled
14.8
21
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like80 d1w2ia_
not modelled
14.7
24
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like81 d1viaa_
not modelled
14.6
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Shikimate kinase (AroK)82 c2r8bA_
not modelled
14.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2452;PDBTitle: the crystal structure of the protein atu2452 of unknown function from2 agrobacterium tumefaciens str. c58
83 c1kk3A_
not modelled
14.4
36
PDB header: translationChain: A: PDB Molecule: eif2gamma;PDBTitle: structure of the wild-type large gamma subunit of2 initiation factor eif2 from pyrococcus abyssi complexed3 with gdp-mg2+
84 c1wb1C_
not modelled
14.4
36
PDB header: protein synthesisChain: C: PDB Molecule: translation elongation factor selb;PDBTitle: crystal structure of translation elongation factor selb2 from methanococcus maripaludis in complex with gdp
85 d1wf3a1
not modelled
14.1
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins86 c3lxwA_
not modelled
13.5
33
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 1;PDBTitle: crystal structure of human gtpase imap family member 1
87 d2gc6a1
not modelled
13.3
16
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: Folylpolyglutamate synthetase, C-terminal domain88 c3a1wA_
not modelled
13.2
14
PDB header: transport proteinChain: A: PDB Molecule: iron(ii) transport protein b;PDBTitle: crystal structue of the g domain of t. maritima feob iron2 iransporter
89 c3lx8A_
not modelled
13.0
30
PDB header: metal transportChain: A: PDB Molecule: ferrous iron uptake transporter protein b;PDBTitle: crystal structure of gdp-bound nfeob from s. thermophilus
90 c2elfA_
not modelled
13.0
27
PDB header: translationChain: A: PDB Molecule: protein translation elongation factor 1a;PDBTitle: crystal structure of the selb-like elongation factor ef-pyl2 from methanosarcina mazei
91 c4proD_
not modelled
12.9
10
PDB header: serine proteaseChain: D: PDB Molecule: alpha-lytic protease;PDBTitle: alpha-lytic protease complexed with pro region
92 c3o93A_
not modelled
12.8
16
PDB header: hydrolaseChain: A: PDB Molecule: nicotinamidase;PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
93 c3dukD_
not modelled
12.7
10
PDB header: unknown functionChain: D: PDB Molecule: ntf2-like protein of unknown function;PDBTitle: crystal structure of a ntf2-like protein of unknown function2 (mfla_0564) from methylobacillus flagellatus kt at 2.200 a resolution
94 c3hynA_
not modelled
12.6
20
PDB header: signaling proteinChain: A: PDB Molecule: putative signal transduction protein;PDBTitle: crystal structure of a putative signal transduction protein2 (eubrec_0645) from eubacterium rectale atcc 33656 at 1.20 a3 resolution
95 c3gkqB_
not modelled
12.6
47
PDB header: oxidoreductaseChain: B: PDB Molecule: terminal oxygenase component of carbazole 1,9a-PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 novosphingobium sp. ka1
96 c1qgeD_
not modelled
12.6
18
PDB header: hydrolaseChain: D: PDB Molecule: protein (triacylglycerol hydrolase);PDBTitle: new crystal form of pseudomonas glumae (formerly chromobacterium2 viscosum atcc 6918) lipase
97 c2wsmB_
not modelled
12.5
29
PDB header: metal binding proteinChain: B: PDB Molecule: hydrogenase expression/formation protein (hypb);PDBTitle: crystal structure of hydrogenase maturation factor hypb from2 archaeoglobus fulgidus
98 d2hh8a1
not modelled
12.5
13
Fold: YdfO-likeSuperfamily: YdfO-likeFamily: YdfO-like99 c3snhA_
not modelled
12.5
27
PDB header: endocytosisChain: A: PDB Molecule: dynamin-1;PDBTitle: crystal structure of nucleotide-free human dynamin1