| 1 | c1gupC_ |
|
 |
100.0 |
100 |
PDB header:nucleotidyltransferase
Chain: C: PDB Molecule:galactose-1-phosphate uridylyltransferase;
PDBTitle: structure of nucleotidyltransferase complexed with udp-2 galactose
|
| 2 | c1zwjA_ |
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative galactose-1-phosphate uridyl transferase;
PDBTitle: x-ray structure of galt-like protein from arabidopsis thaliana2 at5g18200
|
| 3 | d1guqa1 |
|
 |
100.0 |
99 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 4 | d1guqa2 |
|
 |
100.0 |
100 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 5 | d1z84a1 |
|
 |
100.0 |
25 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 6 | d1z84a2 |
|
 |
100.0 |
24 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 7 | c3lb5B_ |
|
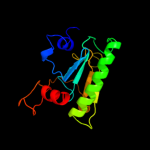 |
99.9 |
19 |
PDB header:cell cycle
Chain: B: PDB Molecule:hit-like protein involved in cell-cycle regulation;
PDBTitle: crystal structure of hit-like protein involved in cell-cycle2 regulation from bartonella henselae with unknown ligand
|
| 8 | c3anoA_ |
|
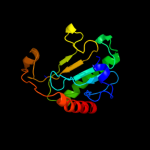 |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:ap-4-a phosphorylase;
PDBTitle: crystal structure of a novel diadenosine 5',5'''-p1,p4-tetraphosphate2 phosphorylase from mycobacterium tuberculosis h37rv
|
| 9 | c3o0mB_ |
|
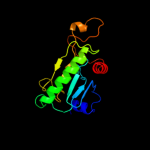 |
99.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family protein;
PDBTitle: crystal structure of a zn-bound histidine triad family protein from2 mycobacterium smegmatis
|
| 10 | c3l7xA_ |
|
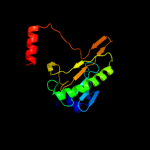 |
99.9 |
19 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative hit-like protein involved in cell-cycle
PDBTitle: the crystal structure of smu.412c from streptococcus mutans ua159
|
| 11 | c3ksvA_ |
|
 |
99.9 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: hypothetical protein from leishmania major
|
| 12 | c3imiB_ |
|
 |
99.9 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hit family protein;
PDBTitle: 2.01 angstrom resolution crystal structure of a hit family protein2 from bacillus anthracis str. 'ames ancestor'
|
| 13 | c3p0tB_ |
|
 |
99.9 |
18 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an hit-like protein from mycobacterium2 paratuberculosis
|
| 14 | d1y23a_ |
|
 |
99.9 |
18 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 15 | c2eo4A_ |
|
 |
99.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:150aa long hypothetical histidine triad nucleotide-binding
PDBTitle: crystal structure of hypothetical histidine triad nucleotide-binding2 protein st2152 from sulfolobus tokodaii strain7
|
| 16 | c3n1tE_ |
|
 |
99.9 |
13 |
PDB header:hydrolase
Chain: E: PDB Molecule:hit-like protein hint;
PDBTitle: crystal structure of the h101a mutant echint gmp complex
|
| 17 | d1xqua_ |
|
 |
99.9 |
19 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 18 | c1xquA_ |
|
 |
99.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:hit family hydrolase;
PDBTitle: hit family hydrolase from clostridium thermocellum cth-393
|
| 19 | d1rzya_ |
|
 |
99.9 |
19 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 20 | d1fita_ |
|
 |
99.9 |
21 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 21 | d1kpfa_ |
|
not modelled |
99.9 |
18 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 22 | d2oika1 |
|
not modelled |
99.9 |
16 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 23 | d1emsa1 |
|
not modelled |
99.9 |
20 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 24 | c3r6fA_ |
|
not modelled |
99.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:hit family protein;
PDBTitle: crystal structure of a zinc-containing hit family protein from2 encephalitozoon cuniculi
|
| 25 | c3oj7A_ |
|
not modelled |
99.8 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative histidine triad family protein;
PDBTitle: crystal structure of a histidine triad family protein from entamoeba2 histolytica, bound to sulfate
|
| 26 | c1emsB_ |
|
not modelled |
99.8 |
19 |
PDB header:antitumor protein
Chain: B: PDB Molecule:nit-fragile histidine triad fusion protein;
PDBTitle: crystal structure of the c. elegans nitfhit protein
|
| 27 | c3i24B_ |
|
not modelled |
99.6 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family hydrolase;
PDBTitle: crystal structure of a hit family hydrolase protein from2 vibrio fischeri. northeast structural genomics consortium3 target id vfr176
|
| 28 | c3nrdB_ |
|
not modelled |
99.6 |
16 |
PDB header:nucleotide binding protein
Chain: B: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
|
| 29 | c3i4sB_ |
|
not modelled |
99.6 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidine triad protein;
PDBTitle: crystal structure of histidine triad protein blr8122 from2 bradyrhizobium japonicum
|
| 30 | c3oheA_ |
|
not modelled |
99.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad protein (maqu_1709) from2 marinobacter aquaeolei vt8 at 1.20 a resolution
|
| 31 | d3bl9a1 |
|
not modelled |
98.7 |
18 |
Fold:HIT-like
Superfamily:HIT-like
Family:mRNA decapping enzyme DcpS C-terminal domain |
| 32 | c3bl9B_ |
|
not modelled |
98.6 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:scavenger mrna-decapping enzyme dcps;
PDBTitle: synthetic gene encoded dcps bound to inhibitor dg157493
|
| 33 | d1vlra1 |
|
not modelled |
98.4 |
21 |
Fold:HIT-like
Superfamily:HIT-like
Family:mRNA decapping enzyme DcpS C-terminal domain |
| 34 | c1xmlA_ |
|
not modelled |
98.4 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock-like protein 1;
PDBTitle: structure of human dcps
|
| 35 | d2pofa1 |
|
not modelled |
85.2 |
14 |
Fold:HIT-like
Superfamily:HIT-like
Family:CDH-like |
| 36 | c1m6vE_ |
|
not modelled |
34.9 |
13 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 37 | d1vpra1 |
|
not modelled |
22.3 |
60 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Dinoflagellate luciferase repeat |
| 38 | c3bddD_ |
|
not modelled |
22.3 |
27 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein marr;
PDBTitle: crystal structure of a putative multiple antibiotic-resistance2 repressor (ssu05_1136) from streptococcus suis 89/1591 at 2.20 a3 resolution
|
| 39 | c3g2bA_ |
|
not modelled |
19.5 |
28 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:coenzyme pqq synthesis protein d;
PDBTitle: crystal structure of pqqd from xanthomonas campestris
|
| 40 | c3ohmB_ |
|
not modelled |
18.2 |
25 |
PDB header:signaling protein / hydrolase
Chain: B: PDB Molecule:1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase
PDBTitle: crystal structure of activated g alpha q bound to its effector2 phospholipase c beta 3
|
| 41 | d1zaka2 |
|
not modelled |
15.2 |
56 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 42 | d1ukfa_ |
|
not modelled |
14.7 |
19 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Avirulence protein Avrpph3 |
| 43 | d1v58a1 |
|
not modelled |
14.0 |
50 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbC/DsbG C-terminal domain-like |
| 44 | c1t3bA_ |
|
not modelled |
13.8 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: x-ray structure of dsbc from haemophilus influenzae
|
| 45 | d1qasa3 |
|
not modelled |
13.5 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 46 | c1v57A_ |
|
not modelled |
13.0 |
50 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbg;
PDBTitle: crystal structure of the disulfide bond isomerase dsbg
|
| 47 | c1jzdA_ |
|
not modelled |
12.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: dsbc-dsbdalpha complex
|
| 48 | d2nvna1 |
|
not modelled |
11.6 |
18 |
Fold:ssDNA-binding transcriptional regulator domain
Superfamily:ssDNA-binding transcriptional regulator domain
Family:PMN2A0962/syc2379c-like |
| 49 | c2rodB_ |
|
not modelled |
10.9 |
24 |
PDB header:apoptosis
Chain: B: PDB Molecule:noxa;
PDBTitle: solution structure of mcl-1 complexed with noxaa
|
| 50 | d1rk4a2 |
|
not modelled |
10.7 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 51 | d2psba1 |
|
not modelled |
10.6 |
33 |
Fold:YerB-like
Superfamily:YerB-like
Family:YerB-like |
| 52 | c2psbA_ |
|
not modelled |
10.6 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yerb protein;
PDBTitle: crystal structure of yerb protein from bacillus subtilis.2 northeast structural genomics target sr586
|
| 53 | d1eeja1 |
|
not modelled |
10.6 |
38 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbC/DsbG C-terminal domain-like |
| 54 | c1hyuA_ |
|
not modelled |
10.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: crystal structure of intact ahpf
|
| 55 | c2hwtA_ |
|
not modelled |
10.4 |
19 |
PDB header:replication, hydrolase
Chain: A: PDB Molecule:putative replicase-associated protein;
PDBTitle: nmr solution structure of the master-rep protein nuclease2 domain (2-95) from the faba bean necrotic yellows virus
|
| 56 | d1ji8a_ |
|
not modelled |
10.4 |
11 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
| 57 | d2v4jc1 |
|
not modelled |
9.7 |
11 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
| 58 | d2drpa2 |
|
not modelled |
9.6 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 59 | c2elpA_ |
|
not modelled |
9.0 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
|
| 60 | d1t3ba1 |
|
not modelled |
8.4 |
38 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbC/DsbG C-terminal domain-like |
| 61 | c3ghaA_ |
|
not modelled |
8.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:disulfide bond formation protein d;
PDBTitle: crystal structure of etda-treated bdbd (reduced)
|
| 62 | c3gv1A_ |
|
not modelled |
8.1 |
63 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:disulfide interchange protein;
PDBTitle: crystal structure of disulfide interchange protein from neisseria2 gonorrhoeae
|
| 63 | d1dqwa_ |
|
not modelled |
8.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 64 | d1ee8a1 |
|
not modelled |
7.9 |
21 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 65 | d1igna2 |
|
not modelled |
7.9 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:DNA-binding domain of rap1 |
| 66 | d1k3sa_ |
|
not modelled |
7.7 |
20 |
Fold:Secretion chaperone-like
Superfamily:Type III secretory system chaperone-like
Family:Type III secretory system chaperone |
| 67 | c3c6cA_ |
|
not modelled |
7.7 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
| 68 | c3lotC_ |
|
not modelled |
7.7 |
36 |
PDB header:structure genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
|
| 69 | c3e49A_ |
|
not modelled |
7.1 |
36 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849 with a tim barrel fold;
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
|
| 70 | c3e02A_ |
|
not modelled |
6.9 |
33 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 71 | c3no5C_ |
|
not modelled |
6.8 |
27 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 72 | d2fbka1 |
|
not modelled |
6.2 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 73 | c3chvA_ |
|
not modelled |
6.2 |
27 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 74 | c3gn3B_ |
|
not modelled |
6.2 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative protein-disulfide isomerase;
PDBTitle: crystal structure of a putative protein-disulfide isomerase from2 pseudomonas syringae to 2.5a resolution.
|
| 75 | d1aisa1 |
|
not modelled |
6.1 |
15 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
| 76 | c3nznA_ |
|
not modelled |
6.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin;
PDBTitle: the crystal structure of the glutaredoxin from methanosarcina mazei2 go1
|
| 77 | c2fd4A_ |
|
not modelled |
6.1 |
25 |
PDB header:ligase
Chain: A: PDB Molecule:avirulence protein avrptob;
PDBTitle: crystal structure of avrptob (436-553)
|
| 78 | c3nqnB_ |
|
not modelled |
6.0 |
47 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein with unknown function. (dr_2006) from2 deinococcus radiodurans at 1.88 a resolution
|
| 79 | c2ab3A_ |
|
not modelled |
5.9 |
45 |
PDB header:rna binding protein
Chain: A: PDB Molecule:znf29;
PDBTitle: solution structures and characterization of hiv rre iib rna2 targeting zinc finger proteins
|
| 80 | d1wika_ |
|
not modelled |
5.9 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 81 | d2csta_ |
|
not modelled |
5.7 |
7 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 82 | c3oopA_ |
|
not modelled |
5.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2960 protein;
PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
|
| 83 | d2fiya1 |
|
not modelled |
5.6 |
22 |
Fold:FdhE-like
Superfamily:FdhE-like
Family:FdhE-like |
| 84 | c2y7eA_ |
|
not modelled |
5.6 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
| 85 | d1dl0a_ |
|
not modelled |
5.5 |
50 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:omega toxin-like
Family:Spider toxins |
| 86 | c1dl0A_ |
|
not modelled |
5.5 |
50 |
PDB header:toxin
Chain: A: PDB Molecule:j-atracotoxin-hv1c;
PDBTitle: solution structure of the insecticidal neurotoxin j-2 atracotoxin-hv1c
|
| 87 | d1fm4a_ |
|
not modelled |
5.5 |
16 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
| 88 | c2hw0A_ |
|
not modelled |
5.4 |
28 |
PDB header:hydrolase, replication
Chain: A: PDB Molecule:replicase;
PDBTitle: nmr solution structure of the nuclease domain from the2 replicator initiator protein from porcine circovirus pcv2
|
| 89 | d1s6la1 |
|
not modelled |
5.4 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MerB N-terminal domain-like |
| 90 | d1qnaa1 |
|
not modelled |
5.4 |
12 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
| 91 | c1zypB_ |
|
not modelled |
5.3 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: synchrotron reduced form of the n-terminal domain of2 salmonella typhimurium ahpf
|
| 92 | d1egoa_ |
|
not modelled |
5.3 |
25 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 93 | d1k0ma2 |
|
not modelled |
5.1 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 94 | d1t3ca_ |
|
not modelled |
5.1 |
25 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Clostridium neurotoxins, catalytic domain |
| 95 | d1vr7a1 |
|
not modelled |
5.1 |
16 |
Fold:S-adenosylmethionine decarboxylase
Superfamily:S-adenosylmethionine decarboxylase
Family:Bacterial S-adenosylmethionine decarboxylase |

