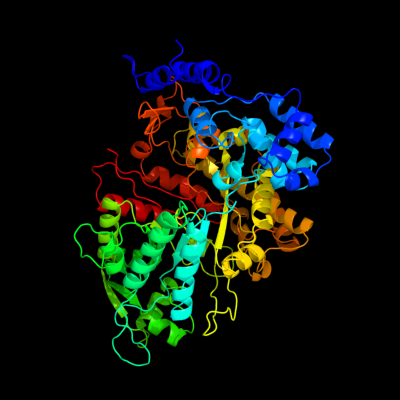
| 1 | c1pemA_
|
|
 |
100.0 |
90 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase 2 alpha
PDBTitle: ribonucleotide reductase protein r1e from salmonella2 typhimurium
|
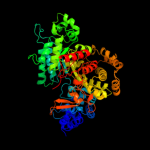
| 2 | c3r1rB_
|
|
 |
100.0 |
23 |
PDB header:complex (oxidoreductase/peptide)
Chain: B: PDB Molecule:ribonucleotide reductase r1 protein;
PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
|
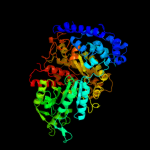
| 3 | c2wghA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large
PDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
|
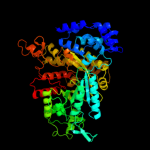
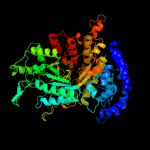
| 4 | c3hnfA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large subunit;
PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
|
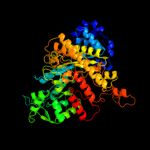
| 5 | c2cvuA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large chain
PDBTitle: structures of yeast ribonucleotide reductase i
|
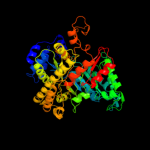
| 6 | c1xjeA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleotide reductase, b12-dependent;
PDBTitle: structural mechanism of allosteric substrate specificity in a2 ribonucleotide reductase: dttp-gdp complex
|
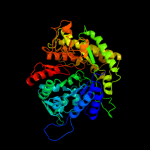
| 7 | d1rlra2
|
|
 |
100.0 |
26 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
| 8 | d1peqa2
|
|
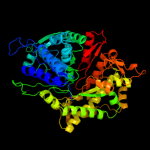 |
100.0 |
91 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
| 9 | d1l1la_
|
|
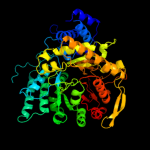 |
100.0 |
19 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:B12-dependent (class II) ribonucleotide reductase |
| 10 | d1peqa1
|
|
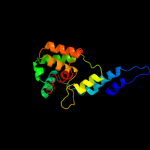 |
100.0 |
85 |
Fold:R1 subunit of ribonucleotide reductase, N-terminal domain
Superfamily:R1 subunit of ribonucleotide reductase, N-terminal domain
Family:R1 subunit of ribonucleotide reductase, N-terminal domain |
| 11 | d1rlra1
|
|
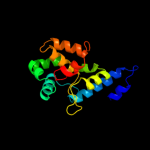 |
100.0 |
14 |
Fold:R1 subunit of ribonucleotide reductase, N-terminal domain
Superfamily:R1 subunit of ribonucleotide reductase, N-terminal domain
Family:R1 subunit of ribonucleotide reductase, N-terminal domain |
| 12 | d1hk8a_
|
|
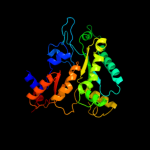 |
95.0 |
17 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
| 13 | c1hk8A_
|
|
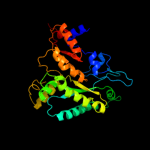 |
95.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
|
| 14 | d1h16a_
|
|
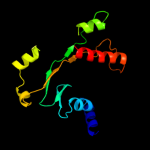 |
76.3 |
14 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 15 | c3pg6D_
|
|
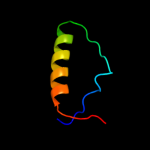 |
53.3 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:e3 ubiquitin-protein ligase dtx3l;
PDBTitle: the carboxyl terminal domain of human deltex 3-like
|
| 16 | d1qhma_
|
|
 |
32.3 |
14 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 17 | d2okqa1
|
|
 |
29.1 |
19 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YbaA-like |
| 18 | d1geqa_
|
|
 |
27.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 19 | d1h3ob_
|
|
 |
22.9 |
13 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 20 | d2ccqa1
|
|
 |
22.4 |
16 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
| 21 | c2okqB_ |
|
not modelled |
20.1 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ybaa;
PDBTitle: crystal structure of unknown conserved ybaa protein from2 shigella flexneri
|
| 22 | d1jlia_ |
|
not modelled |
15.7 |
22 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Short-chain cytokines |
| 23 | d1msza_ |
|
not modelled |
13.8 |
17 |
Fold:IF3-like
Superfamily:R3H domain
Family:R3H domain |
| 24 | c1mszA_ |
|
not modelled |
13.8 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein smubp-2;
PDBTitle: solution structure of the r3h domain from human smubp-2
|
| 25 | d2b4va2 |
|
not modelled |
12.7 |
44 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:RNA editing terminal uridyl transferase 2, RET2, catalytic domain |
| 26 | c3pe0B_ |
|
not modelled |
12.7 |
24 |
PDB header:structural protein
Chain: B: PDB Molecule:plectin;
PDBTitle: structure of the central region of the plakin domain of plectin
|
| 27 | d1v93a_ |
|
not modelled |
12.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Methylenetetrahydrofolate reductase |
| 28 | c1pgjA_ |
|
not modelled |
12.0 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
|
| 29 | d1v74b_ |
|
not modelled |
11.6 |
18 |
Fold:Four-helical up-and-down bundle
Superfamily:Colicin D immunity protein
Family:Colicin D immunity protein |
| 30 | d1vkna2 |
|
not modelled |
10.9 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 31 | c2iz1C_ |
|
not modelled |
10.9 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
| 32 | d1ic8a2 |
|
not modelled |
10.8 |
16 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:POU-specific domain |
| 33 | c3rcqA_ |
|
not modelled |
10.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartyl/asparaginyl beta-hydroxylase;
PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
|
| 34 | c2v3sB_ |
|
not modelled |
10.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:serine/threonine-protein kinase osr1;
PDBTitle: structural insights into the recognition of substrates and2 activators by the osr1 kinase
|
| 35 | d2uyoa1 |
|
not modelled |
10.3 |
14 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:ML2640-like |
| 36 | d1vmha_ |
|
not modelled |
10.2 |
10 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
| 37 | c2f3oB_ |
|
not modelled |
10.1 |
18 |
PDB header:unknown function
Chain: B: PDB Molecule:pyruvate formate-lyase 2;
PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
|
| 38 | d1vmfa_ |
|
not modelled |
9.7 |
10 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
| 39 | c2p4qA_ |
|
not modelled |
9.5 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating 1;
PDBTitle: crystal structure analysis of gnd1 in saccharomyces cerevisiae
|
| 40 | c3fwnB_ |
|
not modelled |
9.2 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
|
| 41 | d1vmja_ |
|
not modelled |
9.2 |
20 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
| 42 | d1j09a2 |
|
not modelled |
8.9 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 43 | d2d5ua1 |
|
not modelled |
8.6 |
16 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
| 44 | c2p6hB_ |
|
not modelled |
8.2 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
|
| 45 | d1r9da_ |
|
not modelled |
7.7 |
17 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 46 | d1gv2a2 |
|
not modelled |
7.7 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 47 | c1pgqA_ |
|
not modelled |
7.6 |
39 |
PDB header:oxidoreductase (choh(d)-nadp+(a))
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: crystallographic study of coenzyme, coenzyme analogue and substrate2 binding in 6-phosphogluconate dehydrogenase: implications for nadp3 specificity and the enzyme mechanism
|
| 48 | c1idzA_ |
|
not modelled |
7.3 |
21 |
PDB header:dna-binding protein
Chain: A: PDB Molecule:mouse c-myb dna-binding domain repeat 3;
PDBTitle: structure of myb transforming protein, nmr, 20 structures
|
| 49 | c2xrgA_ |
|
not modelled |
7.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
| 50 | d1wiga2 |
|
not modelled |
7.2 |
33 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 51 | d2d0ob1 |
|
not modelled |
7.1 |
16 |
Fold:Anticodon-binding domain-like
Superfamily:B12-dependent dehydatase associated subunit
Family:Dehydratase-reactivating factor beta subunit |
| 52 | c2p6cB_ |
|
not modelled |
6.9 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:aq_2013 protein;
PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
|
| 53 | c3ng3A_ |
|
not modelled |
6.9 |
31 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
| 54 | d2csba2 |
|
not modelled |
6.8 |
42 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Topoisomerase V repeat domain |
| 55 | c3hm6X_ |
|
not modelled |
6.8 |
16 |
PDB header:signaling protein
Chain: X: PDB Molecule:plexin-b1;
PDBTitle: crystal structure of the cytoplasmic domain of human plexin b1
|
| 56 | d2b3wa1 |
|
not modelled |
6.6 |
20 |
Fold:YbiA-like
Superfamily:YbiA-like
Family:YbiA-like |
| 57 | c2qlwA_ |
|
not modelled |
6.4 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:rhau;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
|
| 58 | c2qlxA_ |
|
not modelled |
6.4 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:l-rhamnose mutarotase;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
|
| 59 | d1vpha_ |
|
not modelled |
6.3 |
16 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
| 60 | c2c1fA_ |
|
not modelled |
6.3 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:bifunctional endo-1,4-beta-xylanase a;
PDBTitle: the structure of the family 11 xylanase from neocallimastix2 patriciarum
|
| 61 | d2g5ca2 |
|
not modelled |
6.2 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 62 | c3dzbA_ |
|
not modelled |
6.0 |
32 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
|
| 63 | d1f0ya2 |
|
not modelled |
6.0 |
33 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 64 | c3a9lB_ |
|
not modelled |
6.0 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:poly-gamma-glutamate hydrolase;
PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
|
| 65 | c2is9A_ |
|
not modelled |
5.9 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:defective in cullin neddylation protein 1;
PDBTitle: structure of yeast dcn-1
|
| 66 | d1mbja_ |
|
not modelled |
5.8 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 67 | c1mbjA_ |
|
not modelled |
5.8 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:myb proto-oncogene protein;
PDBTitle: mouse c-myb dna-binding domain repeat 3
|
| 68 | c1obfO_ |
|
not modelled |
5.8 |
19 |
PDB header:glycolytic pathway
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.73 resolution.
|
| 69 | d1k3ta2 |
|
not modelled |
5.7 |
22 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 70 | d1jfla1 |
|
not modelled |
5.7 |
32 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 71 | d1h4ga_ |
|
not modelled |
5.6 |
24 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 72 | d1f5ja_ |
|
not modelled |
5.6 |
29 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 73 | c2jr7A_ |
|
not modelled |
5.6 |
26 |
PDB header:metal binding protein
Chain: A: PDB Molecule:dph3 homolog;
PDBTitle: solution structure of human desr1
|
| 74 | c3ivuB_ |
|
not modelled |
5.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 75 | c3iymA_ |
|
not modelled |
5.6 |
20 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein;
PDBTitle: backbone trace of the capsid protein dimer of a fungal partitivirus2 from electron cryomicroscopy and homology modeling
|
| 76 | d1x8da1 |
|
not modelled |
5.5 |
27 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YiiL-like |
| 77 | d1te1b_ |
|
not modelled |
5.4 |
29 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 78 | d1m4wa_ |
|
not modelled |
5.4 |
29 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 79 | d1wgea1 |
|
not modelled |
5.4 |
23 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 80 | d2awia2 |
|
not modelled |
5.4 |
10 |
Fold:alpha-alpha superhelix
Superfamily:TPR-like
Family:PrgX C-terminal domain-like |
| 81 | d1xyna_ |
|
not modelled |
5.4 |
24 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 82 | c2dcjA_ |
|
not modelled |
5.4 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:xylanase j;
PDBTitle: a two-domain structure of alkaliphilic xynj from bacillus sp. 41m-1
|
| 83 | c2g5cD_ |
|
not modelled |
5.2 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 84 | c2kzuA_ |
|
not modelled |
5.2 |
31 |
PDB header:apoptosis
Chain: A: PDB Molecule:death-associated protein 6;
PDBTitle: daxx helical bundle (dhb) domain / rassf1c complex
|
| 85 | d1t6gc_ |
|
not modelled |
5.1 |
18 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 86 | d1xi8a3 |
|
not modelled |
5.1 |
28 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 87 | d1at3a_ |
|
not modelled |
5.1 |
40 |
Fold:Herpes virus serine proteinase, assemblin
Superfamily:Herpes virus serine proteinase, assemblin
Family:Herpes virus serine proteinase, assemblin |
| 88 | d1igoa_ |
|
not modelled |
5.1 |
18 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 89 | c2p6uA_ |
|
not modelled |
5.1 |
28 |
PDB header:dna binding protein
Chain: A: PDB Molecule:afuhel308 helicase;
PDBTitle: apo structure of the hel308 superfamily 2 helicase
|
| 90 | c2pv7B_ |
|
not modelled |
5.0 |
28 |
PDB header:isomerase, oxidoreductase
Chain: B: PDB Molecule:t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)
PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
|