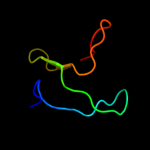
1 c2vv5D_
100.0
18
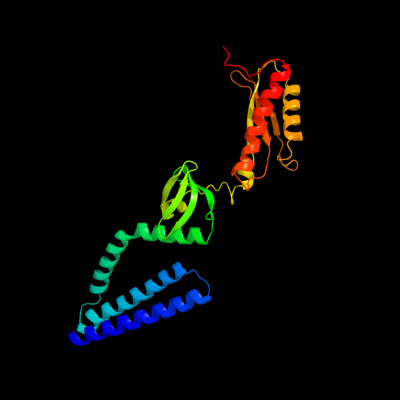
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
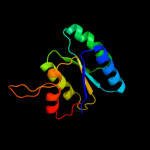
2 d2vv5a1
99.8
23
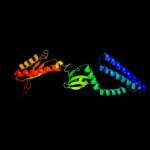
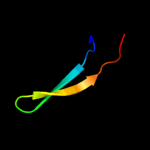
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain3 d2vv5a2
99.2
19
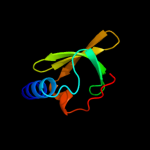
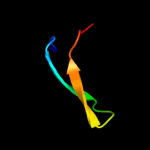
Fold: Ferredoxin-likeSuperfamily: Mechanosensitive channel protein MscS (YggB), C-terminal domainFamily: Mechanosensitive channel protein MscS (YggB), C-terminal domain4 d2vv5a3
98.7
14
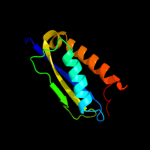
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane regionSuperfamily: Mechanosensitive channel protein MscS (YggB), transmembrane regionFamily: Mechanosensitive channel protein MscS (YggB), transmembrane region5 d1nz9a_
80.7
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain6 d1nppa2
78.9
17
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain7 d2hqha1
78.6
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain8 c2kvqG_
76.8
13
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
9 c2jvvA_
76.8
13
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
10 c2e6zA_
74.6
13
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
11 c2zkrt_
70.4
13
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
12 c2l82A_
69.5
16
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or32;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
13 d1t9ha1
63.1
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like14 c3p8bB_
61.1
30
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
15 d1u0la1
60.0
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like16 c2rcnA_
59.5
19
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
17 d2gvha1
56.7
14
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like18 c2gvhC_
54.3
18
PDB header: hydrolaseChain: C: PDB Molecule: agr_l_2016p;PDBTitle: crystal structure of acyl-coa hydrolase (15159470) from agrobacterium2 tumefaciens at 2.65 a resolution
19 d2eyqa1
53.7
16
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like20 c2yv5A_
52.5
17
PDB header: hydrolaseChain: A: PDB Molecule: yjeq protein;PDBTitle: crystal structure of yjeq from aquifex aeolicus
21 d1whka_
not modelled
52.5
16
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain22 d1whma_
not modelled
51.6
16
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain23 d1y7ua1
not modelled
50.9
25
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like24 c1t9hA_
not modelled
50.7
22
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: the crystal structure of yloq, a circularly permuted gtpase.
25 c3b7kA_
not modelled
50.3
25
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coenzyme a thioesterase 12;PDBTitle: human acyl-coenzyme a thioesterase 12
26 d2gvha2
not modelled
45.3
19
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like27 c1m1gB_
not modelled
44.2
18
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
28 d2coya1
not modelled
43.8
28
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain29 c4a1cS_
not modelled
43.0
18
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
30 d1ylia1
not modelled
42.3
18
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like31 d1vpma_
not modelled
41.1
32
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like32 d2cqaa1
not modelled
38.8
28
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain33 c4a2iV_
not modelled
38.8
21
PDB header: ribosome/hydrolaseChain: V: PDB Molecule: putative ribosome biogenesis gtpase rsga;PDBTitle: cryo-electron microscopy structure of the 30s subunit in complex with2 the yjeq biogenesis factor
34 d1vqot1
not modelled
38.5
32
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e35 c1u0lB_
not modelled
37.0
22
PDB header: hydrolaseChain: B: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of yjeq from thermotoga maritima
36 c2v1oF_
not modelled
35.1
11
PDB header: hydrolaseChain: F: PDB Molecule: cytosolic acyl coenzyme a thioester hydrolase;PDBTitle: crystal structure of n-terminal domain of acyl-coa2 thioesterase 7
37 d2cp6a1
not modelled
35.0
29
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain38 d2do3a1
not modelled
33.9
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like39 d1ppje2
not modelled
33.7
8
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor40 c3iz5Y_
not modelled
31.8
18
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
41 d1yvca1
not modelled
31.4
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain42 d1yeza1
not modelled
30.7
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain43 c2qq2C_
not modelled
30.0
11
PDB header: hydrolaseChain: C: PDB Molecule: cytosolic acyl coenzyme a thioester hydrolase;PDBTitle: crystal structure of c-terminal domain of human acyl-coa thioesterase2 7
44 d2coza1
not modelled
29.7
17
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain45 c2xhcA_
not modelled
29.3
22
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of thermotoga maritima n-utilization substance g2 (nusg)
46 d1bcce2
not modelled
28.6
8
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor47 c2z0wA_
not modelled
27.7
24
PDB header: protein bindingChain: A: PDB Molecule: cap-gly domain-containing linker protein 4;PDBTitle: crystal structure of the 2nd cap-gly domain in human restin-2 like protein 2 reveals a swapped-dimer
48 c3d6lA_
not modelled
27.1
23
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of cj0915, a hexameric hotdog fold2 thioesterase of campylobacter jejuni
49 d2e3ha1
not modelled
26.9
33
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain50 d2cp5a1
not modelled
26.9
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain51 d2cp3a1
not modelled
26.7
33
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain52 c2eisA_
not modelled
26.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein tthb207;PDBTitle: x-ray structure of acyl-coa hydrolase-like protein, tt1379, from2 thermus thermophilus hb8
53 d2e3ia1
not modelled
26.4
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain54 d1whja_
not modelled
25.8
32
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain55 c2e4hA_
not modelled
24.7
30
PDB header: structural proteinChain: A: PDB Molecule: restin;PDBTitle: solution structure of cytoskeletal protein in complex with2 tubulin tail
56 d1ayia_
not modelled
24.6
24
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins57 d2cowa1
not modelled
24.2
22
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain58 d2cp2a1
not modelled
23.8
19
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain59 d2vlqa1
not modelled
23.4
24
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins60 c3owvA_
not modelled
23.3
15
PDB header: hydrolaseChain: A: PDB Molecule: dna-entry nuclease;PDBTitle: structural insights into catalytic and substrate binding mechanisms of2 the strategic enda nuclease from streptococcus pneumoniae
61 c2no8A_
not modelled
22.2
30
PDB header: immune systemChain: A: PDB Molecule: colicin-e2 immunity protein;PDBTitle: nmr structure analysis of the colicin immuntiy protein im2
62 d2cp0a1
not modelled
22.2
22
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain63 c3mayE_
not modelled
20.1
27
PDB header: heme-binding proteinChain: E: PDB Molecule: possible exported protein;PDBTitle: crystal structure of a secreted mycobacterium tuberculosis heme-2 binding protein
64 d1gxha_
not modelled
19.9
30
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins65 c3lwgB_
not modelled
19.4
4
PDB header: unknown functionChain: B: PDB Molecule: hp0420 homologue;PDBTitle: crystal structure of hp0420-homologue c46a from helicobacter2 felis
66 d1njjb1
not modelled
18.8
33
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like67 d1whha_
not modelled
17.4
21
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain68 d2btva_
not modelled
17.2
26
Fold: Reovirus inner layer core protein p3Superfamily: Reovirus inner layer core protein p3Family: Orbivirus core69 c2zqeA_
not modelled
16.4
35
PDB header: dna binding proteinChain: A: PDB Molecule: muts2 protein;PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
70 d2cbpa_
not modelled
16.2
25
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like71 d1i9ga_
not modelled
15.8
8
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like72 d2toda1
not modelled
15.6
33
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like73 d2je6i1
not modelled
15.1
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like74 c2btvB_
not modelled
14.7
26
PDB header: virusChain: B: PDB Molecule: protein (vp3 core protein);PDBTitle: atomic model for bluetongue virus (btv) core
75 d1y4oa1
not modelled
14.3
19
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain76 d2ot2a1
not modelled
13.7
20
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like77 d1sroa_
not modelled
13.7
12
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like78 c1s1iU_
not modelled
13.4
18
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l26-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
79 d1bbua1
not modelled
13.3
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain80 d1ws8a_
not modelled
12.6
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like81 d1e1oa1
not modelled
12.3
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain82 d1e32a3
not modelled
12.2
21
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like83 d2g50a1
not modelled
12.1
14
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain84 d3d3ra1
not modelled
12.1
25
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like85 d1r4va_
not modelled
11.8
23
Fold: Histone-foldSuperfamily: Histone-foldFamily: Bacterial histone-fold protein86 d1g7sa1
not modelled
11.7
26
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors87 c2qlwA_
not modelled
11.6
21
PDB header: isomeraseChain: A: PDB Molecule: rhau;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
88 c2qlxA_
not modelled
11.6
21
PDB header: isomeraseChain: A: PDB Molecule: l-rhamnose mutarotase;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
89 d1wwia1
not modelled
11.5
14
Fold: Histone-foldSuperfamily: Histone-foldFamily: Bacterial histone-fold protein90 d1ubea2
not modelled
11.5
7
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain91 c2gutA_
not modelled
11.5
23
PDB header: transcriptionChain: A: PDB Molecule: arc/mediator, positive cofactor 2 glutamine/q-PDBTitle: solution structure of the trans-activation domain of the2 human co-activator arc105
92 d1mo6a2
not modelled
11.5
7
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain93 c3d3rA_
not modelled
11.4
25
PDB header: chaperoneChain: A: PDB Molecule: hydrogenase assembly chaperone hypc/hupf;PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
94 d1d7ka1
not modelled
11.2
33
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like95 c3g7gG_
not modelled
11.1
18
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: upf0311 protein ca_c3321;PDBTitle: crystal structure of the protein with unknown function from2 clostridium acetobutylicum atcc 824
96 d1g7sa2
not modelled
11.1
19
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors97 d1e0ta1
not modelled
11.1
21
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain98 d1jera_
not modelled
11.0
29
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like99 c1jerA_
not modelled
11.0
29
PDB header: electron transportChain: A: PDB Molecule: cucumber stellacyanin;PDBTitle: cucumber stellacyanin, cu2+, ph 7.0