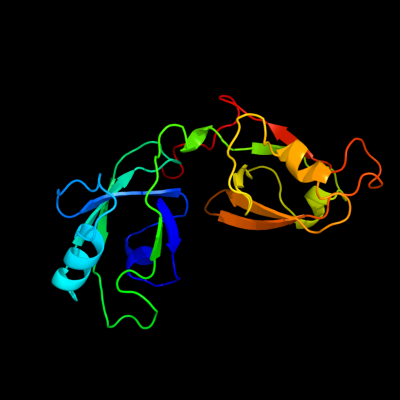
| 1 | c3rleA_
|
|
 |
99.9 |
19 |
PDB header:membrane protein
Chain: A: PDB Molecule:golgi reassembly-stacking protein 2;
PDBTitle: crystal structure of grasp55 grasp domain (residues 7-208)
|
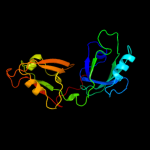
| 2 | c3pv5B_
|
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:degq;
PDBTitle: structure of legionella fallonii degq (n189g/p190g variant)
|
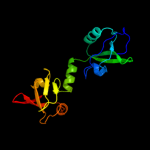
| 3 | c4a8aI_
|
|
 |
99.8 |
18 |
PDB header:hydrolase/hydrolase
Chain: I: PDB Molecule:periplasmic ph-dependent serine endoprotease degq;
PDBTitle: asymmetric cryo-em reconstruction of e. coli degq 12-mer in complex2 with lysozyme
|
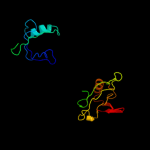
| 4 | c1p1dA_
|
|
 |
99.8 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:glutamate receptor interacting protein;
PDBTitle: structural insights into the inter-domain chaperoning of2 tandem pdz domains in glutamate receptor interacting3 proteins
|
| 5 | c2ka9A_
|
|
 |
99.7 |
14 |
PDB header:cell adhesion
Chain: A: PDB Molecule:disks large homolog 4;
PDBTitle: solution structure of psd-95 pdz12 complexed with cypin2 peptide
|
| 6 | c2qt5A_
|
|
 |
99.7 |
22 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:glutamate receptor-interacting protein 1;
PDBTitle: crystal structure of grip1 pdz12 in complex with the fras12 peptide
|
| 7 | c3b4rB_
|
|
 |
99.7 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative zinc metalloprotease mj0392;
PDBTitle: site-2 protease from methanocaldococcus jannaschii
|
| 8 | c3r0hA_
|
|
 |
99.7 |
14 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:inactivation-no-after-potential d protein;
PDBTitle: structure of inad pdz45 in complex with ng2 peptide
|
| 9 | c1w9qB_
|
|
 |
99.6 |
14 |
PDB header:cell adhesion
Chain: B: PDB Molecule:syntenin 1;
PDBTitle: crystal structure of the pdz tandem of human syntenin in2 complex with tnefaf peptide
|
| 10 | c1u3bA_
|
|
 |
99.6 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:amyloid beta a4 precursor protein-binding,
PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
|
| 11 | c2xkxB_
|
|
 |
99.5 |
14 |
PDB header:structural protein
Chain: B: PDB Molecule:disks large homolog 4;
PDBTitle: single particle analysis of psd-95 in negative stain
|
| 12 | c2p3wB_
|
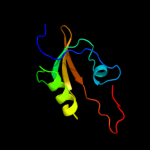
|
 |
99.4 |
31 |
PDB header:protein binding
Chain: B: PDB Molecule:probable serine protease htra3;
PDBTitle: crystal structure of the htra3 pdz domain bound to a phage-derived2 ligand (fgrwv)
|
| 13 | d1lcya1
|
|
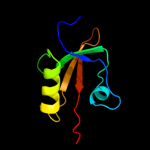 |
99.3 |
29 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 14 | c3i18A_
|
|
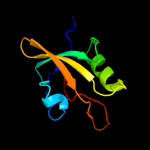 |
99.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo2051 protein;
PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
|
| 15 | c1lcyA_
|
|
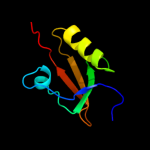 |
99.3 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:htra2 serine protease;
PDBTitle: crystal structure of the mitochondrial serine protease htra2
|
| 16 | d2z9ia1
|
|
 |
99.3 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 17 | c2kl1A_
|
|
 |
99.3 |
28 |
PDB header:protein binding
Chain: A: PDB Molecule:ylbl protein;
PDBTitle: solution structure of gtr34c from geobacillus thermodenitrificans.2 northeast structural genomics consortium target gtr34c
|
| 18 | c2zpmA_
|
|
 |
99.3 |
89 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 19 | c3stjC_
|
|
 |
99.3 |
28 |
PDB header:hydrolase
Chain: C: PDB Molecule:protease degq;
PDBTitle: crystal structure of the protease + pdz1 domain of degq from2 escherichia coli
|
| 20 | c2joaA_
|
|
 |
99.2 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:serine protease htra1;
PDBTitle: htra1 bound to an optimized peptide: nmr assignment of pdz2 domain and ligand resonances
|
| 21 | c3pv4A_ |
|
not modelled |
99.2 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:degq;
PDBTitle: structure of legionella fallonii degq (delta-pdz2 variant)
|
| 22 | c2kjpA_ |
|
not modelled |
99.2 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ylbl;
PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
|
| 23 | c2zplA_ |
|
not modelled |
99.2 |
99 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain a
|
| 24 | d1fc6a3 |
|
not modelled |
99.1 |
24 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:Tail specific protease PDZ domain |
| 25 | d2hgaa1 |
|
not modelled |
99.1 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:MTH1368 C-terminal domain-like |
| 26 | d2i4sa1 |
|
not modelled |
99.1 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:EpsC C-terminal domain-like |
| 27 | c3qo6B_ |
|
not modelled |
99.1 |
25 |
PDB header:photosynthesis
Chain: B: PDB Molecule:protease do-like 1, chloroplastic;
PDBTitle: crystal structure analysis of the plant protease deg1
|
| 28 | d1ky9b2 |
|
not modelled |
99.0 |
27 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 29 | d2i6va1 |
|
not modelled |
99.0 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:EpsC C-terminal domain-like |
| 30 | c3gdsA_ |
|
not modelled |
99.0 |
23 |
PDB header:hydrolase/hydrolase activator
Chain: A: PDB Molecule:protease degs;
PDBTitle: crystal structure of degs h198p/d320a mutant modified by dfp in2 complex with dnrdgnvyyf peptide
|
| 31 | d1ky9a1 |
|
not modelled |
98.9 |
31 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 32 | c1z87A_ |
|
not modelled |
98.9 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:alpha-1-syntrophin;
PDBTitle: solution structure of the split ph-pdz supramodule of alpha-2 syntrophin
|
| 33 | c2eaqA_ |
|
not modelled |
98.9 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:lim domain only protein 7;
PDBTitle: crystal structure of pdz domain of kiaa0858 (lim), ms07932 from homo sapiens
|
| 34 | d1w9ea1 |
|
not modelled |
98.9 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 35 | c1ky9A_ |
|
not modelled |
98.9 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:protease do;
PDBTitle: crystal structure of degp (htra)
|
| 36 | d1qaua_ |
|
not modelled |
98.9 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 37 | d1x5qa1 |
|
not modelled |
98.8 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 38 | c2krgA_ |
|
not modelled |
98.8 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf1;
PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
|
| 39 | c2kjdA_ |
|
not modelled |
98.8 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:sodium/hydrogen exchange regulatory cofactor nhe-
PDBTitle: solution structure of extended pdz2 domain from nherf1 (150-2 270)
|
| 40 | c3diwB_ |
|
not modelled |
98.8 |
24 |
PDB header:signaling protein/cell adhesion
Chain: B: PDB Molecule:tax1-binding protein 3;
PDBTitle: c-terminal beta-catenin bound tip-1 structure
|
| 41 | d1sota1 |
|
not modelled |
98.8 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 42 | d1ozia_ |
|
not modelled |
98.8 |
21 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 43 | d1wh1a_ |
|
not modelled |
98.8 |
24 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 44 | d1m5za_ |
|
not modelled |
98.8 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 45 | d1wifa_ |
|
not modelled |
98.8 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 46 | c2vsvB_ |
|
not modelled |
98.8 |
18 |
PDB header:protein-binding
Chain: B: PDB Molecule:rhophilin-2;
PDBTitle: crystal structure of the pdz domain of human rhophilin-2
|
| 47 | c3eggC_ |
|
not modelled |
98.8 |
30 |
PDB header:hydrolase
Chain: C: PDB Molecule:spinophilin;
PDBTitle: crystal structure of a complex between protein phosphatase 1 alpha2 (pp1) and the pp1 binding and pdz domains of spinophilin
|
| 48 | c2vwrA_ |
|
not modelled |
98.8 |
17 |
PDB header:protein-binding
Chain: A: PDB Molecule:ligand of numb protein x 2;
PDBTitle: crystal structure of the second pdz domain of numb-binding2 protein 2
|
| 49 | c2he4A_ |
|
not modelled |
98.8 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf2;
PDBTitle: the crystal structure of the second pdz domain of human2 nherf-2 (slc9a3r2) interacting with a mode 1 pdz binding3 motif
|
| 50 | d1wfga_ |
|
not modelled |
98.7 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 51 | d1p1da2 |
|
not modelled |
98.7 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 52 | d1q3oa_ |
|
not modelled |
98.7 |
31 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 53 | c3l4fD_ |
|
not modelled |
98.7 |
25 |
PDB header:signaling protein/protein binding
Chain: D: PDB Molecule:sh3 and multiple ankyrin repeat domains protein
PDBTitle: crystal structure of betapix coiled-coil domain and shank2 pdz complex
|
| 54 | c3shuB_ |
|
not modelled |
98.7 |
27 |
PDB header:cell adhesion
Chain: B: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of zo-1 pdz3
|
| 55 | d1pdra_ |
|
not modelled |
98.7 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 56 | c2eehA_ |
|
not modelled |
98.7 |
30 |
PDB header:metal binding protein
Chain: A: PDB Molecule:pdz domain-containing protein 7;
PDBTitle: solution structure of first pdz domain of pdz domain2 containing protein 7
|
| 57 | c2komA_ |
|
not modelled |
98.7 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:partitioning defective 3 homolog;
PDBTitle: solution structure of humar par-3b pdz2 (residues 451-549)
|
| 58 | d1ueqa_ |
|
not modelled |
98.7 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 59 | d1rgwa_ |
|
not modelled |
98.7 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 60 | d1uf1a_ |
|
not modelled |
98.7 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 61 | c2egkC_ |
|
not modelled |
98.7 |
23 |
PDB header:protein binding
Chain: C: PDB Molecule:general receptor for phosphoinositides 1-
PDBTitle: crystal structure of tamalin pdz-intrinsic ligand fusion2 protein
|
| 62 | c2q3gA_ |
|
not modelled |
98.7 |
21 |
PDB header:structural genomics
Chain: A: PDB Molecule:pdz and lim domain protein 7;
PDBTitle: structure of the pdz domain of human pdlim7 bound to a c-2 terminal extension from human beta-tropomyosin
|
| 63 | d1q7xa_ |
|
not modelled |
98.7 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 64 | d1g9oa_ |
|
not modelled |
98.7 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 65 | c2iwnA_ |
|
not modelled |
98.7 |
27 |
PDB header:signaling protein
Chain: A: PDB Molecule:multiple pdz domain protein;
PDBTitle: 3rd pdz domain of multiple pdz domain protein mpdz (casp2 target)
|
| 66 | d2f5ya1 |
|
not modelled |
98.7 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 67 | d1d5ga_ |
|
not modelled |
98.7 |
29 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 68 | d2f0aa1 |
|
not modelled |
98.7 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 69 | d1uhpa_ |
|
not modelled |
98.7 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 70 | d1tp5a1 |
|
not modelled |
98.7 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 71 | d1be9a_ |
|
not modelled |
98.7 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 72 | c2gzvA_ |
|
not modelled |
98.7 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:prkca-binding protein;
PDBTitle: the cystal structure of the pdz domain of human pick1 (casp target)
|
| 73 | c2jxoA_ |
|
not modelled |
98.7 |
25 |
PDB header:protein binding
Chain: A: PDB Molecule:ezrin-radixin-moesin-binding phosphoprotein 50;
PDBTitle: structure of the second pdz domain of nherf-1
|
| 74 | c3khfA_ |
|
not modelled |
98.6 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:microtubule-associated serine/threonine-protein
PDBTitle: the crystal structure of the pdz domain of human microtubule2 associated serine/threonine kinase 3 (mast3)
|
| 75 | c2jilA_ |
|
not modelled |
98.6 |
27 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate receptor interacting protein-1;
PDBTitle: crystal structure of 2nd pdz domain of glutamate receptor2 interacting protein-1 (grip1)
|
| 76 | d1t2ma1 |
|
not modelled |
98.6 |
28 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 77 | c2v90E_ |
|
not modelled |
98.6 |
21 |
PDB header:protein-binding
Chain: E: PDB Molecule:pdz domain-containing protein 3;
PDBTitle: crystal structure of the 3rd pdz domain of intestine- and2 kidney-enriched pdz domain ikepp (pdzd3)
|
| 78 | c3k1rA_ |
|
not modelled |
98.6 |
20 |
PDB header:structural protein
Chain: A: PDB Molecule:harmonin;
PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
|
| 79 | d2fe5a1 |
|
not modelled |
98.6 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 80 | c2qktB_ |
|
not modelled |
98.6 |
20 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:inactivation-no-after-potential d protein;
PDBTitle: crystal structure of the 5th pdz domain of inad
|
| 81 | c3qikA_ |
|
not modelled |
98.6 |
19 |
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phosphatidylinositol 3,4,5-trisphosphate-dependent rac
PDBTitle: crystal structure of the first pdz domain of prex1
|
| 82 | c2e7kA_ |
|
not modelled |
98.6 |
24 |
PDB header:membrane protein
Chain: A: PDB Molecule:maguk p55 subfamily member 2;
PDBTitle: solution structure of the pdz domain from human maguk p552 subfamily member 2
|
| 83 | c2ogpA_ |
|
not modelled |
98.6 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:partitioning-defective 3 homolog;
PDBTitle: solution structure of the second pdz domain of par-3
|
| 84 | c2k1zA_ |
|
not modelled |
98.6 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:partitioning-defective 3 homolog;
PDBTitle: solution structure of par-3 pdz3
|
| 85 | c2jikB_ |
|
not modelled |
98.6 |
26 |
PDB header:membrane protein
Chain: B: PDB Molecule:synaptojanin-2 binding protein;
PDBTitle: crystal structure of pdz domain of synaptojanin-2 binding2 protein
|
| 86 | d1va8a1 |
|
not modelled |
98.6 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 87 | c2jreA_ |
|
not modelled |
98.6 |
30 |
PDB header:de novo protein
Chain: A: PDB Molecule:c60-1 pdz domain peptide;
PDBTitle: c60-1, a pdz domain designed using statistical coupling2 analysis
|
| 88 | d1ihja_ |
|
not modelled |
98.6 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 89 | c2d92A_ |
|
not modelled |
98.6 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:inad-like protein;
PDBTitle: solution structure of the fifth pdz domain of inad-like2 protein
|
| 90 | c2fneB_ |
|
not modelled |
98.6 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:multiple pdz domain protein;
PDBTitle: the crystal structure of the 13th pdz domain of mpdz
|
| 91 | c2dkrA_ |
|
not modelled |
98.6 |
19 |
PDB header:protein transport
Chain: A: PDB Molecule:lin-7 homolog b;
PDBTitle: solution structure of the pdz domain from human lin-72 homolog b
|
| 92 | c2edzA_ |
|
not modelled |
98.6 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:pdz domain-containing protein 1;
PDBTitle: solution structures of the pdz domain of mus musculus pdz2 domain-containing protein 1
|
| 93 | d1ry4a_ |
|
not modelled |
98.6 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 94 | c2o2tB_ |
|
not modelled |
98.6 |
16 |
PDB header:structural protein
Chain: B: PDB Molecule:multiple pdz domain protein;
PDBTitle: the crystal structure of the 1st pdz domain of mpdz
|
| 95 | d1wjla_ |
|
not modelled |
98.6 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 96 | c2omjA_ |
|
not modelled |
98.6 |
29 |
PDB header:cell adhesion
Chain: A: PDB Molecule:rho guanine nucleotide exchange factor 12;
PDBTitle: solution structure of larg pdz domain
|
| 97 | c2qg1A_ |
|
not modelled |
98.6 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:multiple pdz domain protein;
PDBTitle: crystal structure of the 11th pdz domain of mpdz (mupp1)
|
| 98 | c2d90A_ |
|
not modelled |
98.6 |
19 |
PDB header:protein binding
Chain: A: PDB Molecule:pdz domain containing protein 1;
PDBTitle: solution structure of the third pdz domain of pdz domain2 containing protein 1
|
| 99 | c3qglD_ |
|
not modelled |
98.6 |
32 |
PDB header:protein binding
Chain: D: PDB Molecule:sorting nexin-27;
PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
|
| 100 | d1whaa_ |
|
not modelled |
98.6 |
24 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 101 | c2dmzA_ |
|
not modelled |
98.6 |
33 |
PDB header:protein binding
Chain: A: PDB Molecule:inad-like protein;
PDBTitle: solution structure of the third pdz domain of human inad-2 like protein
|
| 102 | c2dluA_ |
|
not modelled |
98.6 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:inad-like protein;
PDBTitle: solution structure of the second pdz domain of human inad-2 like protein
|
| 103 | d2byga1 |
|
not modelled |
98.6 |
21 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 104 | d1kwaa_ |
|
not modelled |
98.6 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 105 | d2cssa1 |
|
not modelled |
98.6 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 106 | c3hpmA_ |
|
not modelled |
98.6 |
23 |
PDB header:protein binding
Chain: A: PDB Molecule:protein interacting with prkca 1;
PDBTitle: oxidized dimeric pick1 pdz c46g mutant in complex with the carboxyl2 tail peptide of glur2
|
| 107 | d1p1da1 |
|
not modelled |
98.6 |
32 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 108 | d1wf8a1 |
|
not modelled |
98.6 |
32 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 109 | d1whda_ |
|
not modelled |
98.6 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 110 | c2i04B_ |
|
not modelled |
98.6 |
21 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:membrane-associated guanylate kinase, ww and pdz
PDBTitle: x-ray crystal structure of magi-1 pdz1 bound to the c-2 terminal peptide of hpv18 e6
|
| 111 | c2z17A_ |
|
not modelled |
98.6 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:pleckstrin homology sec7 and coiled-coil domains-
PDBTitle: crystal sturcture of pdz domain from human pleckstrin2 homology, sec7
|
| 112 | c2v1wB_ |
|
not modelled |
98.6 |
21 |
PDB header:structural protein
Chain: B: PDB Molecule:pdz and lim domain protein 4;
PDBTitle: crystal structure of human lim protein ril (pdlim4) pdz2 domain bound to the c-terminal peptide of human alpha-3 actinin-1
|
| 113 | d1n7ea_ |
|
not modelled |
98.6 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 114 | d1um1a_ |
|
not modelled |
98.6 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 115 | c2dm8A_ |
|
not modelled |
98.6 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:inad-like protein;
PDBTitle: solution structure of the eighth pdz domain of human inad-2 like protein
|
| 116 | d1ujda_ |
|
not modelled |
98.6 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 117 | d1wf7a_ |
|
not modelled |
98.6 |
16 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 118 | c3shwA_ |
|
not modelled |
98.6 |
28 |
PDB header:cell adhesion
Chain: A: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of zo-1 pdz3-sh3-guk supramodule complex with2 connexin-45 peptide
|
| 119 | d1vaea_ |
|
not modelled |
98.6 |
21 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 120 | c3b76A_ |
|
not modelled |
98.6 |
28 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase lnx;
PDBTitle: crystal structure of the third pdz domain of human ligand-of-numb2 protein-x (lnx1) in complex with the c-terminal peptide from the3 coxsackievirus and adenovirus receptor
|