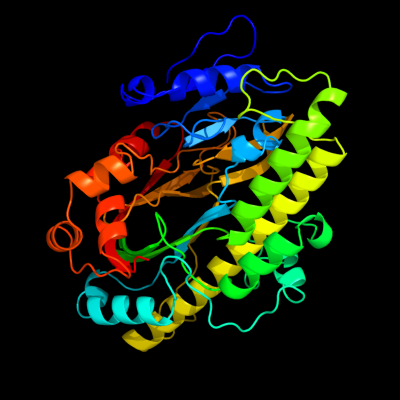
| 1 | d1uc2a_
|
|
 |
100.0 |
29 |
Fold:Hypothetical protein PH1602
Superfamily:Hypothetical protein PH1602
Family:Hypothetical protein PH1602 |
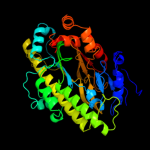
| 2 | c2epgB_
|
|
 |
100.0 |
31 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ttha1785;
PDBTitle: crystal structure of ttha1785
|
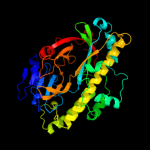
| 3 | d1el6a_
|
|
 |
56.3 |
26 |
Fold:Baseplate structural protein gp11
Superfamily:Baseplate structural protein gp11
Family:Baseplate structural protein gp11 |
| 4 | d1xmta_
|
|
 |
52.7 |
19 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 5 | d2pd4a1
|
|
 |
47.7 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 6 | c2btwA_
|
|
 |
43.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:alr0975 protein;
PDBTitle: crystal structure of alr0975
|
| 7 | d1fxkc_
|
|
 |
35.0 |
16 |
Fold:Long alpha-hairpin
Superfamily:Prefoldin
Family:Prefoldin |
| 8 | c3emkA_
|
|
 |
34.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose/ribitol dehydrogenase;
PDBTitle: 2.5a crystal structure of glucose/ribitol dehydrogenase2 from brucella melitensis
|
| 9 | d1hxha_
|
|
 |
31.9 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 10 | d2bu3a1
|
|
 |
29.4 |
19 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Phytochelatin synthase |
| 11 | c3grkE_
|
|
 |
27.4 |
16 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
| 12 | c2jjyD_
|
|
 |
26.7 |
8 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of francisella tularensis enoyl reductase2 (ftfabi) with bound nad
|
| 13 | c2zdiC_
|
|
 |
25.6 |
27 |
PDB header:chaperone
Chain: C: PDB Molecule:prefoldin subunit alpha;
PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
|
| 14 | c3ek2D_
|
|
 |
25.4 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase2 from burkholderia pseudomallei 1719b
|
| 15 | c2c4rL_
|
|
 |
24.9 |
33 |
PDB header:hydrolase
Chain: L: PDB Molecule:ribonuclease e;
PDBTitle: catalytic domain of e. coli rnase e
|
| 16 | d2h7ma1
|
|
 |
24.5 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 17 | c2bpbB_
|
|
 |
20.5 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sulfite\:cytochrome c oxidoreductase subunit b;
PDBTitle: sulfite dehydrogenase from starkeya novella
|
| 18 | c2o2sA_
|
|
 |
20.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: the structure of t. gondii enoyl acyl carrier protein reductase in2 complex with nad and triclosan
|
| 19 | d1jmxa1
|
|
 |
20.1 |
11 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 20 | c2quoA_
|
|
 |
19.6 |
33 |
PDB header:toxin
Chain: A: PDB Molecule:heat-labile enterotoxin b chain;
PDBTitle: crystal structure of c terminal fragment of clostridium2 perfringens enterotoxin
|
| 21 | d1pbya1 |
|
not modelled |
19.5 |
19 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 22 | d1w96a2 |
|
not modelled |
19.5 |
43 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 23 | d2j9ga2 |
|
not modelled |
18.9 |
27 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 24 | c1jmuE_ |
|
not modelled |
18.6 |
67 |
PDB header:viral protein
Chain: E: PDB Molecule:protein mu-1;
PDBTitle: crystal structure of the reovirus mu1/sigma3 complex
|
| 25 | d1ulza2 |
|
not modelled |
18.4 |
43 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 26 | d1kx7a_ |
|
not modelled |
17.9 |
27 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 27 | c2ptgA_ |
|
not modelled |
17.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: crystal structure of eimeria tenella enoyl reductase
|
| 28 | d1uoua3 |
|
not modelled |
17.7 |
7 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
| 29 | c2ig6B_ |
|
not modelled |
17.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nimc/nima family protein;
PDBTitle: crystal structure of a nimc/nima family protein (ca_c2569) from2 clostridium acetobutylicum at 1.80 a resolution
|
| 30 | c1w4zA_ |
|
not modelled |
17.0 |
20 |
PDB header:antibiotic biosynthesis
Chain: A: PDB Molecule:ketoacyl reductase;
PDBTitle: structure of actinorhodin polyketide (actiii) reductase
|
| 31 | c3gdfA_ |
|
not modelled |
16.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nadp-dependent mannitol dehydrogenase;
PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
|
| 32 | c2nq8B_ |
|
not modelled |
16.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: malarial enoyl acyl acp reductase bound with inh-nad adduct
|
| 33 | c2foiB_ |
|
not modelled |
16.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: synthesis, biological activity, and x-ray crystal structural analysis2 of diaryl ether inhibitors of malarial enoyl acp reductase.
|
| 34 | c3svtA_ |
|
not modelled |
16.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: structure of a short-chain type dehydrogenase/reductase from2 mycobacterium ulcerans
|
| 35 | d1xq1a_ |
|
not modelled |
15.8 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 36 | d1wkqa_ |
|
not modelled |
15.8 |
41 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
| 37 | c3cu4A_ |
|
not modelled |
15.7 |
27 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome c family protein;
PDBTitle: omcf, outer membrance cytochrome f from geobacter2 sulfurreducens
|
| 38 | d2rhca1 |
|
not modelled |
15.7 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 39 | d1r0ua_ |
|
not modelled |
15.3 |
24 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Hypothetical protein YwiB |
| 40 | d1ulua_ |
|
not modelled |
15.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 41 | d1h5qa_ |
|
not modelled |
14.2 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 42 | d1uh5a_ |
|
not modelled |
13.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 43 | d2ag5a1 |
|
not modelled |
13.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 44 | c3giwA_ |
|
not modelled |
12.9 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:protein of unknown function duf574;
PDBTitle: crystal structure of a duf574 family protein (sav_2177) from2 streptomyces avermitilis ma-4680 at 1.45 a resolution
|
| 45 | c3db0B_ |
|
not modelled |
12.8 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lin2891 protein;
PDBTitle: crystal structure of putative pyridoxamine 5'-phosphate oxidase2 (np_472219.1) from listeria innocua at 2.00 a resolution
|
| 46 | c2d0sA_ |
|
not modelled |
12.6 |
17 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome c;
PDBTitle: crystal structure of the cytochrome c552 from moderate2 thermophilic bacterium, hydrogenophilus thermoluteolus
|
| 47 | c2v07A_ |
|
not modelled |
12.3 |
27 |
PDB header:photosynthesis
Chain: A: PDB Molecule:cytochrome c6;
PDBTitle: structure of the arabidopsis thaliana cytochrome c6a v52q2 variant
|
| 48 | c3gr6A_ |
|
not modelled |
12.3 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of the staphylococcus aureus enoyl-acyl2 carrier protein reductase (fabi) in complex with nadp and3 triclosan
|
| 49 | c3ku1E_ |
|
not modelled |
12.0 |
40 |
PDB header:transferase
Chain: E: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of streptococcus pneumoniae sp1610, a2 putative trna (m1a22) methyltransferase, in complex with s-3 adenosyl-l-methionine
|
| 50 | d2tpta3 |
|
not modelled |
11.6 |
3 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
| 51 | c2p91A_ |
|
not modelled |
11.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of enoyl-[acyl-carrier-protein] reductase (nadh)2 from aquifex aeolicus vf5
|
| 52 | d1cc5a_ |
|
not modelled |
11.4 |
36 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 53 | d1qsga_ |
|
not modelled |
11.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 54 | c3ckkA_ |
|
not modelled |
10.8 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(7)-)-methyltransferase;
PDBTitle: crystal structure of human methyltransferase-like protein 1
|
| 55 | d1mz4a_ |
|
not modelled |
10.7 |
21 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 56 | c3qivA_ |
|
not modelled |
10.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase or 3-oxoacyl-[acyl-carrier-
PDBTitle: crystal structure of a putative short-chain dehydrogenase or 3-2 oxoacyl-[acyl-carrier-protein] reductase from mycobacterium3 paratuberculosis atcc baa-968 / k-10
|
| 57 | d1fcdc1 |
|
not modelled |
10.5 |
33 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Two-domain cytochrome c |
| 58 | d1tt2a_ |
|
not modelled |
10.4 |
10 |
Fold:OB-fold
Superfamily:Staphylococcal nuclease
Family:Staphylococcal nuclease |
| 59 | d1cyja_ |
|
not modelled |
10.4 |
27 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 60 | d1brwa3 |
|
not modelled |
10.4 |
13 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
| 61 | c3g5tA_ |
|
not modelled |
10.4 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 3-methyltransferase;
PDBTitle: crystal structure of trans-aconitate 3-methyltransferase2 from yeast
|
| 62 | d1wvec1 |
|
not modelled |
10.4 |
18 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 63 | d2fd5a2 |
|
not modelled |
10.3 |
20 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
| 64 | c2bh1Y_ |
|
not modelled |
10.1 |
23 |
PDB header:transport protein
Chain: Y: PDB Molecule:general secretion pathway protein e,;
PDBTitle: x-ray structure of the general secretion pathway complex of2 the n-terminal domain of epse and the cytosolic domain of3 epsl of vibrio cholerae
|
| 65 | d2bh1x1 |
|
not modelled |
10.1 |
23 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:EspE N-terminal domain-like
Family:GSPII protein E N-terminal domain-like |
| 66 | c3dliB_ |
|
not modelled |
10.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of a sam dependent methyltransferase from2 archaeoglobus fulgidus
|
| 67 | c3bkwB_ |
|
not modelled |
9.8 |
80 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine dependent methyltransferase;
PDBTitle: crystal structure of s-adenosylmethionine dependent methyltransferase2 (np_104914.1) from mesorhizobium loti at 1.60 a resolution
|
| 68 | c1j6qA_ |
|
not modelled |
9.7 |
26 |
PDB header:chaperone
Chain: A: PDB Molecule:cytochrome c maturation protein e;
PDBTitle: solution structure and characterization of the heme2 chaperone ccme
|
| 69 | d1j6qa_ |
|
not modelled |
9.7 |
26 |
Fold:OB-fold
Superfamily:Heme chaperone CcmE
Family:Heme chaperone CcmE |
| 70 | c3lf2B_ |
|
not modelled |
9.6 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain oxidoreductase q9hya2;
PDBTitle: nadph bound structure of the short chain oxidoreductase q9hya2 from2 pseudomonas aeruginosa pao1 containing an atypical catalytic center
|
| 71 | d1ja9a_ |
|
not modelled |
9.5 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | c3fzgA_ |
|
not modelled |
9.4 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:16s rrna methylase;
PDBTitle: structure of the 16s rrna methylase arma
|
| 73 | c3dlcA_ |
|
not modelled |
9.4 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:putative s-adenosyl-l-methionine-dependent
PDBTitle: crystal structure of a putative s-adenosyl-l-methionine-dependent2 methyltransferase (mmp1179) from methanococcus maripaludis at 1.15 a3 resolution
|
| 74 | d1vz6a_ |
|
not modelled |
9.4 |
12 |
Fold:DmpA/ArgJ-like
Superfamily:DmpA/ArgJ-like
Family:ArgJ-like |
| 75 | d2h00a1 |
|
not modelled |
9.2 |
45 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Methyltransferase 10 domain |
| 76 | d1xxla_ |
|
not modelled |
9.2 |
80 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 77 | c2p35A_ |
|
not modelled |
9.1 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 2-methyltransferase;
PDBTitle: crystal structure of trans-aconitate methyltransferase from2 agrobacterium tumefaciens
|
| 78 | c2qioA_ |
|
not modelled |
9.0 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 79 | c2p8jA_ |
|
not modelled |
9.0 |
60 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine-dependent methyltransferase;
PDBTitle: crystal structure of s-adenosylmethionine-dependent methyltransferase2 (np_349143.1) from clostridium acetobutylicum at 2.00 a resolution
|
| 80 | c3dxyA_ |
|
not modelled |
9.0 |
60 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(7)-)-methyltransferase;
PDBTitle: crystal structure of ectrmb in complex with sam
|
| 81 | d2avna1 |
|
not modelled |
9.0 |
60 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:UbiE/COQ5-like |
| 82 | c3i9fB_ |
|
not modelled |
9.0 |
80 |
PDB header:transferase
Chain: B: PDB Molecule:putative type 11 methyltransferase;
PDBTitle: crystal structure of a putative type 11 methyltransferase2 from sulfolobus solfataricus
|
| 83 | c3l8dA_ |
|
not modelled |
9.0 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of methyltransferase from bacillus2 thuringiensis
|
| 84 | c3k31B_ |
|
not modelled |
8.9 |
4 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of eonyl-(acyl-carrier-protein) reductase from2 anaplasma phagocytophilum in complex with nad at 1.9a resolution
|
| 85 | c3d2lC_ |
|
not modelled |
8.8 |
100 |
PDB header:transferase
Chain: C: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of sam-dependent methyltransferase (zp_00538691.1)2 from exiguobacterium sp. 255-15 at 1.90 a resolution
|
| 86 | d1wfwa_ |
|
not modelled |
8.7 |
20 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 87 | c2hxgB_ |
|
not modelled |
8.7 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:l-arabinose isomerase;
PDBTitle: crystal structure of mn2+ bound ecai
|
| 88 | c3merA_ |
|
not modelled |
8.7 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:slr1183 protein;
PDBTitle: crystal structure of the methyltransferase slr1183 from2 synechocystis sp. pcc 6803, northeast structural genomics3 consortium target sgr145
|
| 89 | c3opnA_ |
|
not modelled |
8.7 |
60 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative hemolysin;
PDBTitle: the crystal structure of a putative hemolysin from lactococcus lactis
|
| 90 | d2fk8a1 |
|
not modelled |
8.6 |
100 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 91 | d1kpga_ |
|
not modelled |
8.6 |
80 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 92 | c2fk8A_ |
|
not modelled |
8.6 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:methoxy mycolic acid synthase 4;
PDBTitle: crystal structure of hma (mmaa4) from mycobacterium tuberculosis2 complexed with s-adenosylmethionine
|
| 93 | d1ctja_ |
|
not modelled |
8.6 |
27 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 94 | c2x5sB_ |
|
not modelled |
8.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:mannose-1-phosphate guanylyltransferase;
PDBTitle: crystal structure of t. maritima gdp-mannose2 pyrophosphorylase in apo state.
|
| 95 | c3egeA_ |
|
not modelled |
8.5 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase from antibiotic biosynthesis
PDBTitle: crystal structure of putative methyltransferase from antibiotic2 biosynthesis pathway (yp_324569.1) from anabaena variabilis atcc3 29413 at 2.40 a resolution
|
| 96 | c3h2bB_ |
|
not modelled |
8.4 |
60 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of the sam-dependent methyltransferase2 cg3271 from corynebacterium glutamicum in complex with s-3 adenosyl-l-homocysteine and pyrophosphate. northeast4 structural genomics consortium target cgr113a
|
| 97 | c3g07C_ |
|
not modelled |
8.3 |
60 |
PDB header:transferase
Chain: C: PDB Molecule:7sk snrna methylphosphate capping enzyme;
PDBTitle: methyltransferase domain of human bicoid-interacting protein2 3 homolog (drosophila)
|
| 98 | c3tl3A_ |
|
not modelled |
8.3 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: structure of a short-chain type dehydrogenase/reductase from2 mycobacterium ulcerans
|
| 99 | d2fcta1 |
|
not modelled |
8.3 |
32 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |