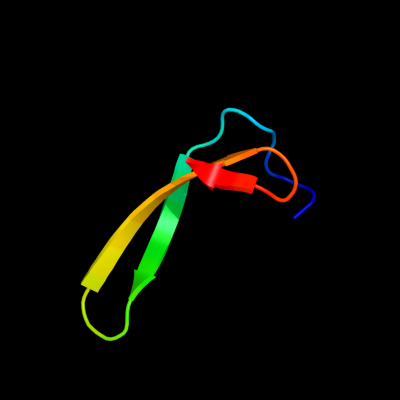
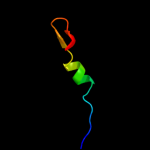
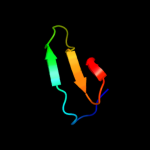
1 c2kytA_
31.5
14
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
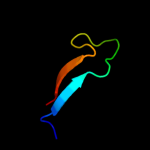
2 c2pqhA_
26.7
20
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: structure of sh3 chimera with a type ii ligand linked to the chain c-2 terminal
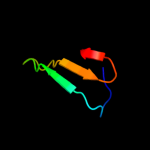
3 d1pwta_
25.4
20
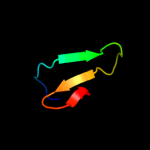
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain4 c3cqtA_
24.6
16
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fyn;PDBTitle: n53i v55l mutant of fyn sh3 domain
5 c2l55A_
24.2
7
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
6 d1gria2
23.8
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain7 c2jttD_
22.2
20
PDB header: calcium binding protein/antitumor proteiChain: D: PDB Molecule: calcyclin-binding protein;PDBTitle: solution structure of calcium loaded s100a6 bound to c-2 terminal siah-1 interacting protein
8 c2nwmA_
21.5
20
PDB header: cell adhesionChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the first sh3 domain of human vinexin2 and its interaction with the peptides from vinculin
9 c3bu2B_
20.6
20
PDB header: rna binding proteinChain: B: PDB Molecule: putative trna-binding protein;PDBTitle: crystal structure of a trna-binding protein from2 staphylococcus saprophyticus subsp. saprophyticus.3 northeast structural genomics consortium target syr77
10 c1negA_
20.6
14
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: crystal structure analysis of n-and c-terminal labeled sh3-2 domain of alpha-chicken spectrin
11 d1nega_
20.6
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain12 c2ed0A_
20.5
11
PDB header: signaling proteinChain: A: PDB Molecule: abl interactor 2;PDBTitle: solution structure of the sh3 domain of abl interactor 22 (abelson interactor 2)
13 d1zata2
20.5
26
Fold: L,D-transpeptidase pre-catalytic domain-likeSuperfamily: L,D-transpeptidase pre-catalytic domain-likeFamily: L,D-transpeptidase pre-catalytic domain-like14 c1zeqX_
20.4
5
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
15 d1uufa1
19.0
11
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain16 c2yunA_
18.4
21
PDB header: protein transportChain: A: PDB Molecule: nostrin;PDBTitle: solution structure of the sh3 domain of human nostrin
17 c2xmfA_
18.0
29
PDB header: motor proteinChain: A: PDB Molecule: myosin 1e sh3;PDBTitle: myosin 1e sh3
18 d1e6ga_
17.9
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain19 d1e6ha_
17.8
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain20 c2drmB_
17.8
14
PDB header: contractile proteinChain: B: PDB Molecule: acanthamoeba myosin ib;PDBTitle: acanthamoeba myosin i sh3 domain bound to acan125
21 d1y7na1
not modelled
16.8
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain22 c2e8gB_
not modelled
16.5
20
PDB header: rna binding proteinChain: B: PDB Molecule: hypothetical protein ph0536;PDBTitle: the structure of protein from p. horikoshii at 1.7 angstrom2 resolution
23 d1oeba_
not modelled
16.5
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain24 c1zx6A_
not modelled
16.4
11
PDB header: protein bindingChain: A: PDB Molecule: ypr154wp;PDBTitle: high-resolution crystal structure of yeast pin3 sh3 domain
25 c2krnA_
not modelled
16.4
11
PDB header: signaling proteinChain: A: PDB Molecule: cd2-associated protein;PDBTitle: high resolution structure of the second sh3 domain of cd2ap
26 c3anuA_
not modelled
16.4
10
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
27 d1wh2a_
not modelled
16.3
20
Fold: GYF/BRK domain-likeSuperfamily: GYF domainFamily: GYF domain28 c2dl4A_
not modelled
16.1
14
PDB header: signaling proteinChain: A: PDB Molecule: protein stac;PDBTitle: solution structure of the first sh3 domain of stac protein
29 d1gria1
not modelled
15.8
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain30 d1eu3a1
not modelled
15.7
50
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain31 d2ux9a1
not modelled
15.7
33
Fold: Dodecin subunit-likeSuperfamily: Dodecin-likeFamily: Dodecin-like32 d1u06a1
not modelled
15.7
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain33 d1i7na1
not modelled
15.7
47
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Synapsin domain34 c3bboL_
not modelled
15.3
18
PDB header: ribosomeChain: L: PDB Molecule: ribosomal protein l13;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
35 c2ekhA_
not modelled
15.1
25
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structures of the sh3 domain of human kiaa0418
36 d1ijwc_
not modelled
14.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain37 c3cf5G_
not modelled
14.7
18
PDB header: ribosome/antibioticChain: G: PDB Molecule: 50s ribosomal protein l13;PDBTitle: thiopeptide antibiotic thiostrepton bound to the large ribosomal2 subunit of deinococcus radiodurans
38 d2zjrg1
not modelled
14.7
18
Fold: Ribosomal protein L13Superfamily: Ribosomal protein L13Family: Ribosomal protein L1339 c2zpmA_
not modelled
14.5
6
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
40 c2vxaL_
not modelled
14.5
27
PDB header: flavoproteinChain: L: PDB Molecule: dodecin;PDBTitle: h.halophila dodecin in complex with riboflavin
41 d1h8ka_
not modelled
14.3
23
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain42 c2d1xD_
not modelled
14.3
11
PDB header: cell invasionChain: D: PDB Molecule: cortactin isoform a;PDBTitle: the crystal structure of the cortactin-sh3 domain and amap1-2 peptide complex
43 c3oqtP_
not modelled
14.2
13
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
44 d1ng2a1
not modelled
14.0
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain45 d1k4us_
not modelled
13.8
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain46 c1uijA_
not modelled
13.7
18
PDB header: sugar binding proteinChain: A: PDB Molecule: beta subunit of beta conglycinin;PDBTitle: crystal structure of soybean beta-conglycinin beta2 homotrimer (i122m/k124w)
47 d2g1la1
not modelled
13.4
11
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain48 d1pk8a1
not modelled
13.3
42
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Synapsin domain49 c3d5bN_
not modelled
13.1
24
PDB header: ribosomeChain: N: PDB Molecule: 50s ribosomal protein l13;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
50 d1qu5a_
not modelled
12.9
12
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain51 c1ov3A_
not modelled
12.9
15
PDB header: oxidoreductase activatorChain: A: PDB Molecule: neutrophil cytosol factor 1;PDBTitle: structure of the p22phox-p47phox complex
52 d1wixa_
not modelled
12.5
9
Fold: CH domain-likeSuperfamily: Hook domainFamily: Hook domain53 c2dbmA_
not modelled
12.0
11
PDB header: transferase, signaling proteinChain: A: PDB Molecule: sh3-containing grb2-like protein 2;PDBTitle: solution structures of the sh3 domain of human sh3-2 containing grb2-like protein 2
54 c2eqiA_
not modelled
12.0
14
PDB header: immune system, hydrolaseChain: A: PDB Molecule: phospholipase c, gamma 2;PDBTitle: solution structure of the sh3 domain from phospholipase c,2 gamma 2
55 c1x2pA_
not modelled
12.0
18
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 2;PDBTitle: solution structure of the sh3 domain of the protein2 arginine n-methyltransferase 2
56 c1xniI_
not modelled
12.0
21
PDB header: cell cycleChain: I: PDB Molecule: tumor suppressor p53-binding protein 1;PDBTitle: tandem tudor domain of 53bp1
57 d2j01n1
not modelled
11.9
24
Fold: Ribosomal protein L13Superfamily: Ribosomal protein L13Family: Ribosomal protein L1358 c2ciuA_
not modelled
11.8
17
PDB header: protein transportChain: A: PDB Molecule: import inner membrane translocase subunit tim21PDBTitle: structure of the ims domain of the mitochondrial import2 protein tim21 from s. cerevisiae
59 c2y6xA_
not modelled
11.7
20
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kd protein;PDBTitle: structure of psb27 from thermosynechococcus elongatus
60 c1u39A_
not modelled
11.6
14
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding,PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
61 c3onrI_
not modelled
11.6
38
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
62 d1hcra_
not modelled
11.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain63 c2o2oA_
not modelled
11.4
14
PDB header: protein bindingChain: A: PDB Molecule: sh3-domain kinase-binding protein 1;PDBTitle: solution structure of domain b from human cin85 protein
64 c2jmcA_
not modelled
11.4
17
PDB header: signaling proteinChain: A: PDB Molecule: spectrin alpha chain, brain and p41 peptidePDBTitle: chimer between spc-sh3 and p41
65 c2dilA_
not modelled
11.3
14
PDB header: cell adhesionChain: A: PDB Molecule: proline-serine-threonine phosphatase-interactingPDBTitle: solution structure of the sh3 domain of the human proline-2 serine-threonine phosphatase-interacting protein 1
66 c2dmoA_
not modelled
11.2
18
PDB header: signaling proteinChain: A: PDB Molecule: neutrophil cytosol factor 2;PDBTitle: refined solution structure of the 1st sh3 domain from human2 neutrophil cytosol factor 2 (ncf-2)
67 c2cucA_
not modelled
11.2
9
PDB header: signaling proteinChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: solution structure of the sh3 domain of the mouse2 hypothetical protein sh3rf2
68 d1uuea_
not modelled
10.9
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain69 d1tuca_
not modelled
10.6
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain70 c2bz8B_
not modelled
10.6
11
PDB header: sh3 domainChain: B: PDB Molecule: sh3-domain kinase binding protein 1;PDBTitle: n-terminal sh3 domain of cin85 bound to cbl-b peptide
71 c1zlmA_
not modelled
10.6
15
PDB header: signaling proteinChain: A: PDB Molecule: osteoclast stimulating factor 1;PDBTitle: crystal structure of the sh3 domain of human osteoclast2 stimulating factor
72 c3i18A_
not modelled
10.6
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2051 protein;PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
73 d1wlpb2
not modelled
10.6
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain74 c1g7tA_
not modelled
10.5
16
PDB header: translationChain: A: PDB Molecule: translation initiation factor if2/eif5b;PDBTitle: x-ray structure of translation initiation factor if2/eif5b2 complexed with gdpnp
75 d1ujya_
not modelled
10.4
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain76 d1wlpb1
not modelled
10.2
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain77 c2kmfA_
not modelled
10.1
17
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kda protein;PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
78 c3ehkC_
not modelled
10.1
15
PDB header: plant proteinChain: C: PDB Molecule: prunin;PDBTitle: crystal structure of pru du amandin, an allergenic protein2 from prunus dulcis
79 c1ma0B_
not modelled
10.1
15
PDB header: oxidoreductaseChain: B: PDB Molecule: glutathione-dependent formaldehyde dehydrogenase;PDBTitle: ternary complex of human glutathione-dependent formaldehyde2 dehydrogenase with nad+ and dodecanoic acid
80 c1wyxA_
not modelled
10.0
14
PDB header: cell adhesionChain: A: PDB Molecule: crk-associated substrate;PDBTitle: the crystal structure of the p130cas sh3 domain at 1.1 a2 resolution
81 d1sema_
not modelled
10.0
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain82 d1ycsb2
not modelled
9.7
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain83 c1q0vA_
not modelled
9.7
13
PDB header: transport bindingChain: A: PDB Molecule: hydrophilic protein; has cysteine rich putativePDBTitle: solution structure of tandem uims of vps27
84 c1f8fA_
not modelled
9.6
19
PDB header: oxidoreductaseChain: A: PDB Molecule: benzyl alcohol dehydrogenase;PDBTitle: crystal structure of benzyl alcohol dehydrogenase from acinetobacter2 calcoaceticus
85 d1ov3a2
not modelled
9.5
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain86 c2epdA_
not modelled
9.4
18
PDB header: protein bindingChain: A: PDB Molecule: rho gtpase-activating protein 4;PDBTitle: solution structure of sh3 domain in rho-gtpase-activating2 protein 4
87 c2l0aA_
not modelled
9.4
11
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 1;PDBTitle: solution nmr structure of signal transducing adapter molecule 1 stam-12 from homo sapiens, northeast structural genomics consortium target3 hr4479e
88 c3a9lB_
not modelled
9.2
10
PDB header: hydrolaseChain: B: PDB Molecule: poly-gamma-glutamate hydrolase;PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
89 d1yvca1
not modelled
9.2
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain90 d1shfa_
not modelled
9.1
22
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain91 d1uj0a_
not modelled
9.1
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain92 c2djqA_
not modelled
9.0
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: the solution structure of the first sh3 domain of mouse sh32 domain containing ring finger 2
93 c3fmaD_
not modelled
9.0
25
PDB header: protein bindingChain: D: PDB Molecule: protein smy2;PDBTitle: crystal structure of the gyf domain of smy2 in complex with a proline-2 rich peptide from bbp/scsf1
94 d1ov3a1
not modelled
8.9
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain95 c1z4hA_
not modelled
8.9
5
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: tor inhibition protein;PDBTitle: the response regulator tori belongs to a new family of2 atypical excisionase
96 d2gych1
not modelled
8.9
12
Fold: Ribosomal protein L13Superfamily: Ribosomal protein L13Family: Ribosomal protein L1397 c2egaA_
not modelled
8.8
14
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the first sh3 domain from human2 kiaa0418 protein
98 d1uhfa_
not modelled
8.7
19
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain99 c3cosD_
not modelled
8.7
23
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase 4;PDBTitle: crystal structure of human class ii alcohol dehydrogenase (adh4) in2 complex with nad and zn