1 c2zvyB_
100.0
35
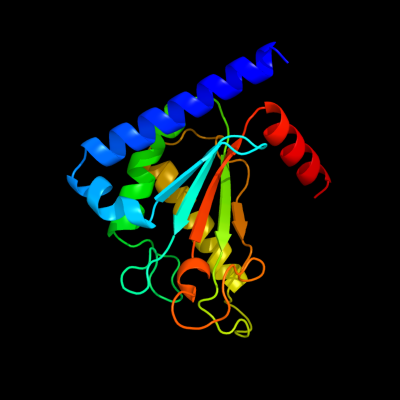
PDB header: membrane proteinChain: B: PDB Molecule: chemotaxis protein motb;PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form ii)
2 c3khnB_
100.0
19
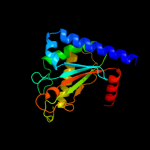
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: motb protein, putative;PDBTitle: crystal structure of putative motb like protein dvu_22282 from desulfovibrio vulgaris.
3 c2l26A_
100.0
23
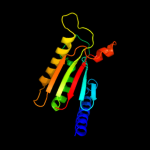
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein rv0899/mt0922;PDBTitle: rv0899 from mycobacterium tuberculosis contains two separated domains
4 c2k1sA_
100.0
21
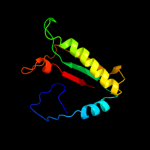
PDB header: lipoproteinChain: A: PDB Molecule: inner membrane lipoprotein yiad;PDBTitle: solution nmr structure of the folded c-terminal fragment of yiad from2 escherichia coli. northeast structural genomics consortium target3 er553.
5 c3cyqM_
100.0
20
PDB header: membrane proteinChain: M: PDB Molecule: chemotaxis protein motb;PDBTitle: the crystal structure of the complex of the c-terminal domain of2 helicobacter pylori motb (residues 125-256) with n-acetylmuramic acid
6 c2zovA_
99.9
34
PDB header: membrane proteinChain: A: PDB Molecule: chemotaxis protein motb;PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form i)
7 c2kgwA_
99.9
24
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: solution structure of the carboxy-terminal domain of ompatb, a pore2 forming protein from mycobacterium tuberculosis
8 d2aizp1
99.9
19
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like9 c3td4D_
99.9
21
PDB header: membrane protein,peptide binding proteinChain: D: PDB Molecule: outer membrane protein omp38;PDBTitle: crystal structure of ompa-like domain from acinetobacter baumannii in2 complex with diaminopimelate
10 d2hqsc1
99.9
22
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like11 d1r1ma_
99.9
20
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like12 c1r1mA_
99.9
20
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein class 4;PDBTitle: structure of the ompa-like domain of rmpm from neisseria2 meningitidis
13 c3ldtA_
99.9
16
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein, ompa family protein;PDBTitle: crystal structure of an outer membrane protein(ompa)from2 legionella pneumophila
14 c3oonA_
99.9
23
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein (tpn50);PDBTitle: the structure of an outer membrance protein from borrelia burgdorferi2 b31
15 c2zf8A_
99.6
27
PDB header: structural proteinChain: A: PDB Molecule: component of sodium-driven polar flagellar motor;PDBTitle: crystal structure of moty
16 d2cz4a1
39.0
12
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein17 d2axla1
31.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like18 c3l7pA_
30.8
13
PDB header: transcriptionChain: A: PDB Molecule: putative nitrogen regulatory protein pii;PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
19 c3latB_
29.9
11
PDB header: hydrolaseChain: B: PDB Molecule: bifunctional autolysin;PDBTitle: crystal structure of staphylococcus peptidoglycan hydrolase2 amie
20 c3o8wA_
29.0
16
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen regulatory protein p-ii (glnb-1);PDBTitle: archaeoglobus fulgidus glnk1
21 d1qy7a_
not modelled
28.7
14
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein22 c2rd5D_
not modelled
27.9
16
PDB header: protein bindingChain: D: PDB Molecule: pii protein;PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
23 c3mhyC_
not modelled
26.5
11
PDB header: signaling proteinChain: C: PDB Molecule: pii-like protein pz;PDBTitle: a new pii protein structure
24 c3bzqA_
not modelled
23.5
19
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: nitrogen regulatory protein p-ii;PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
25 d2piia_
not modelled
21.7
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein26 c2j9dG_
not modelled
21.5
17
PDB header: membrane transportChain: G: PDB Molecule: hypothetical nitrogen regulatory pii-likePDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
27 d1hwua_
not modelled
21.5
21
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein28 d1ul3a_
not modelled
20.7
19
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein29 c3ncpD_
not modelled
20.4
21
PDB header: signaling proteinChain: D: PDB Molecule: nitrogen regulatory protein p-ii (glnb-2);PDBTitle: glnk2 from archaeoglobus fulgidus
30 d2ns1b1
not modelled
20.0
14
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein31 d1z2la1
not modelled
18.7
19
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases32 d1e32a3
not modelled
18.2
16
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like33 d1rbli_
not modelled
16.4
16
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit34 d1h4pa_
not modelled
16.1
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases35 d1svdm1
not modelled
14.9
8
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit36 d1uzhc1
not modelled
14.8
8
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit37 d1e5ma2
not modelled
13.8
9
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related38 d1ir1s_
not modelled
13.4
28
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit39 d1xhja_
not modelled
13.2
8
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like40 c2oghA_
not modelled
13.2
4
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor eif-1;PDBTitle: solution structure of yeast eif1
41 d1vfja_
not modelled
13.1
5
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein42 d1jx4a1
not modelled
13.0
16
Fold: Lesion bypass DNA polymerase (Y-family), little finger domainSuperfamily: Lesion bypass DNA polymerase (Y-family), little finger domainFamily: Lesion bypass DNA polymerase (Y-family), little finger domain43 d1k1sa1
not modelled
13.0
16
Fold: Lesion bypass DNA polymerase (Y-family), little finger domainSuperfamily: Lesion bypass DNA polymerase (Y-family), little finger domainFamily: Lesion bypass DNA polymerase (Y-family), little finger domain44 d1bxni_
not modelled
12.3
20
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit45 d2if1a_
not modelled
12.0
22
Fold: eIF1-likeSuperfamily: eIF1-likeFamily: eIF1-like46 c2qw5B_
not modelled
11.7
8
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
47 c3icgD_
not modelled
11.5
12
PDB header: hydrolaseChain: D: PDB Molecule: endoglucanase d;PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
48 d2gfva2
not modelled
11.4
17
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related49 d1bwvs_
not modelled
11.4
20
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit50 c3o0dF_
not modelled
11.3
25
PDB header: hydrolaseChain: F: PDB Molecule: triacylglycerol lipase;PDBTitle: crystal structure of lip2 lipase from yarrowia lipolytica at 1.7 a2 resolution
51 d1u5ta2
not modelled
11.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain52 c3r8kB_
not modelled
10.7
22
PDB header: apoptosisChain: B: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul protein (hexagonal form)
53 d2gova1
not modelled
10.2
21
Fold: Probable bacterial effector-binding domainSuperfamily: Probable bacterial effector-binding domainFamily: SOUL heme-binding protein54 d1tqya2
not modelled
10.1
12
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related55 c2q3vB_
not modelled
9.9
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein at2g34160;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g34160
56 d1d4oa_
not modelled
9.8
12
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)57 d1qnaa1
not modelled
9.8
14
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain58 d1saza1
not modelled
9.8
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like59 c1ir6A_
not modelled
9.8
15
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease recj;PDBTitle: crystal structure of exonuclease recj bound to manganese
60 d1ir6a_
not modelled
9.8
15
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Exonuclease RecJ61 c2cfuA_
not modelled
9.7
10
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
62 d2ix4a2
not modelled
9.5
18
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related63 c1pt9B_
not modelled
9.4
12
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p) transhydrogenase, mitochondrial;PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
64 c3g7nA_
not modelled
9.1
17
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of a triacylglycerol lipase from2 penicillium expansum at 1.3
65 d1jqna_
not modelled
9.0
15
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase66 d1xdpa4
not modelled
8.5
17
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain67 c3ds8A_
not modelled
8.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2722 protein;PDBTitle: the crysatl structure of the gene lin2722 products from listeria2 innocua
68 c1amuB_
not modelled
7.8
18
PDB header: peptide synthetaseChain: B: PDB Molecule: gramicidin synthetase 1;PDBTitle: phenylalanine activating domain of gramicidin synthetase 12 in a complex with amp and phenylalanine
69 c1nauA_
not modelled
7.6
18
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon;PDBTitle: nmr solution structure of the glucagon antagonist [deshis1,2 desphe6, glu9]glucagon amide in the presence of3 perdeuterated dodecylphosphocholine micelles
70 c1jqoA_
not modelled
7.4
13
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
71 d1jqoa_
not modelled
7.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase72 d2o8ra4
not modelled
7.4
27
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain73 c2rodB_
not modelled
7.2
22
PDB header: apoptosisChain: B: PDB Molecule: noxa;PDBTitle: solution structure of mcl-1 complexed with noxaa
74 c3hluA_
not modelled
7.2
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein duf2179;PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 duf2179 from eubacterium ventriosum
75 d3tgla_
not modelled
7.2
26
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases76 d2j5pa1
not modelled
7.2
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like77 d1ox0a2
not modelled
7.0
20
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related78 c3mmpC_
not modelled
7.0
21
PDB header: transferaseChain: C: PDB Molecule: elongation factor tu 2, elongation factor ts;PDBTitle: structure of the qb replicase, an rna-dependent rna polymerase2 consisting of viral and host proteins
79 d1r3na1
not modelled
7.0
11
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases80 d1tkja1
not modelled
6.9
18
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases81 c1ydoC_
not modelled
6.8
8
PDB header: lyaseChain: C: PDB Molecule: hmg-coa lyase;PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
82 d2v7fa1
not modelled
6.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rps19E-like83 c3aysA_
not modelled
6.5
10
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: gh5 endoglucanase from a ruminal fungus in complex with cellotriose
84 d1ybha1
not modelled
6.4
9
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain85 c2cw6B_
not modelled
6.1
17
PDB header: lyaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa lyase, mitochondrial;PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
86 d2ve8a1
not modelled
6.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like87 c4a1dH_
not modelled
6.0
22
PDB header: ribosomeChain: H: PDB Molecule: rpl35a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
88 d1j3na2
not modelled
5.8
22
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related89 d1e5da2
not modelled
5.8
17
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: ROO N-terminal domain-like90 d1nh2a1
not modelled
5.8
12
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain91 d1amua_
not modelled
5.8
18
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like92 d1uc8a1
not modelled
5.7
42
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Lysine biosynthesis enzyme LysX, N-terminal domain93 c3lubE_
not modelled
5.6
13
PDB header: hydrolaseChain: E: PDB Molecule: putative creatinine amidohydrolase;PDBTitle: crystal structure of putative creatinine amidohydrolase2 (yp_211512.1) from bacteroides fragilis nctc 9343 at 2.11 a3 resolution
94 d1fyba1
not modelled
5.5
18
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors95 c3bzjA_
not modelled
5.3
11
PDB header: hydrolaseChain: A: PDB Molecule: uv endonuclease;PDBTitle: uvde k229l
96 d1nh2a2
not modelled
5.2
15
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain97 c3eyiB_
not modelled
5.2
29
PDB header: dna binding protein/dnaChain: B: PDB Molecule: z-dna-binding protein 1;PDBTitle: the crystal structure of the second z-dna binding domain of2 human dai (zbp1) in complex with z-dna
98 d1v8da_
not modelled
5.1
12
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Hypothetical protein TT1679