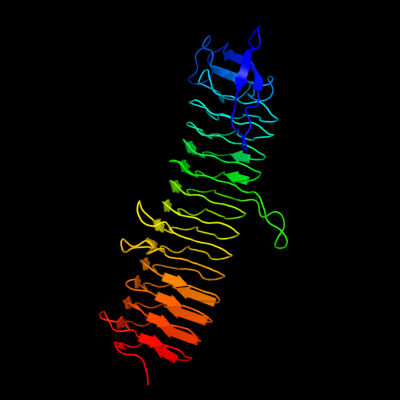
| 1 | d2f9ca1
|
|
 |
100.0 |
65 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:YdcK-like |
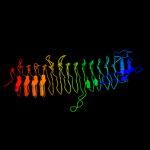
| 2 | c3pmoA_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;
PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
|
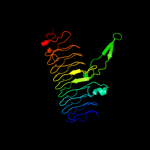
| 3 | c2iu9C_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine
PDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
|
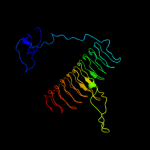
| 4 | c3eh0C_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-
PDBTitle: crystal structure of lpxd from escherichia coli
|
| 5 | c3i3aC_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
|
| 6 | d2jf2a1
|
|
 |
99.9 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
| 7 | d1j2za_
|
|
 |
99.9 |
14 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
| 8 | c3r0sA_
|
|
 |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
|
| 9 | c3fsbB_
|
|
 |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:qdtc;
PDBTitle: crystal structure of qdtc, the dtdp-3-amino-3,6-dideoxy-d-2 glucose n-acetyl transferase from thermoanaerobacterium3 thermosaccharolyticum in complex with coa and dtdp-3-amino-4 quinovose
|
| 10 | c3c8vA_
|
|
 |
99.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_390128.1) from2 desulfovibrio desulfuricans g20 at 2.28 a resolution
|
| 11 | c2oi6A_
|
|
 |
99.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e. coli glmu- complex with udp-glcnac, coa and glcn-1-po4
|
| 12 | c1hm8A_
|
|
 |
99.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine-1-phosphate uridyltransferase;
PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
|
| 13 | c2v0hA_
|
|
 |
99.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
|
| 14 | d2oi6a1
|
|
 |
99.6 |
15 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 15 | d3bswa1
|
|
 |
99.6 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:PglD-like |
| 16 | d1g97a1
|
|
 |
99.6 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 17 | c3srtB_
|
|
 |
99.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:maltose o-acetyltransferase;
PDBTitle: the crystal structure of a maltose o-acetyltransferase from2 clostridium difficile 630
|
| 18 | c3eg4A_
|
|
 |
99.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase from brucella melitensis3 biovar abortus 2308
|
| 19 | c3cj8B_
|
|
 |
99.5 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate n-2 succinyltransferase from enterococcus faecalis v583
|
| 20 | c3mqhD_
|
|
 |
99.5 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:lipopolysaccharides biosynthesis acetyltransferase;
PDBTitle: crystal structure of the 3-n-acetyl transferase wlbb from bordetella2 petrii in complex with coa and udp-3-amino-2-acetamido-2,3-dideoxy3 glucuronic acid
|
| 21 | c3jqyB_ |
|
not modelled |
99.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystal strucutre of the polysia specific acetyltransferase neuo
|
| 22 | c3fttA_ |
|
not modelled |
99.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase sacol2570;
PDBTitle: crystal structure of the galactoside o-acetyltransferase2 from staphylococcus aureus
|
| 23 | c2ic7A_ |
|
not modelled |
99.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:maltose transacetylase;
PDBTitle: crystal structure of maltose transacetylase from2 geobacillus kaustophilus
|
| 24 | d1ocxa_ |
|
not modelled |
99.4 |
15 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 25 | c3ectA_ |
|
not modelled |
99.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide-repeat containing-acetyltransferase;
PDBTitle: crystal structure of the hexapeptide-repeat containing-2 acetyltransferase vca0836 from vibrio cholerae
|
| 26 | d1krra_ |
|
not modelled |
99.4 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 27 | c1t3dB_ |
|
not modelled |
99.4 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
|
| 28 | d1t3da_ |
|
not modelled |
99.4 |
23 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 29 | d1ssqa_ |
|
not modelled |
99.4 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 30 | d3tdta_ |
|
not modelled |
99.3 |
11 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Tetrahydrodipicolinate-N-succinlytransferase, THDP-succinlytransferase, DapD |
| 31 | c2wlgA_ |
|
not modelled |
99.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
|
| 32 | d1qrea_ |
|
not modelled |
99.3 |
11 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 33 | c1qreA_ |
|
not modelled |
99.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: a closer look at the active site of gamma-carbonic anhydrases: high2 resolution crystallographic studies of the carbonic anhydrase from3 methanosarcina thermophila
|
| 34 | d1mr7a_ |
|
not modelled |
99.3 |
11 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 35 | c3mc4A_ |
|
not modelled |
99.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:ww/rsp5/wwp domain:bacterial transferase
PDBTitle: crystal structure of ww/rsp5/wwp domain: bacterial2 transferase hexapeptide repeat: serine o-acetyltransferase3 from brucella melitensis
|
| 36 | c3q1xA_ |
|
not modelled |
99.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of entamoeba histolytica serine acetyltransferase 12 in complex with l-serine
|
| 37 | c3f1xA_ |
|
not modelled |
99.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
| 38 | c3ixcA_ |
|
not modelled |
99.2 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide transferase family protein;
PDBTitle: crystal structure of hexapeptide transferase family protein from2 anaplasma phagocytophilum
|
| 39 | c3r3rA_ |
|
not modelled |
99.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:ferripyochelin binding protein;
PDBTitle: structure of the yrda ferripyochelin binding protein from salmonella2 enterica
|
| 40 | d1xata_ |
|
not modelled |
99.1 |
9 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 41 | c2ggqA_ |
|
not modelled |
99.1 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:401aa long hypothetical glucose-1-phosphate
PDBTitle: complex of hypothetical glucose-1-phosphate thymidylyltransferase from2 sulfolobus tokodaii
|
| 42 | c3eevC_ |
|
not modelled |
99.1 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
|
| 43 | d1v3wa_ |
|
not modelled |
99.1 |
12 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 44 | d1xhda_ |
|
not modelled |
99.0 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 45 | c3kwdA_ |
|
not modelled |
99.0 |
13 |
PDB header:lyase, protein binding, photosynthesis
Chain: A: PDB Molecule:carbon dioxide concentrating mechanism protein;
PDBTitle: inactive truncation of the beta-carboxysomal gamma-carbonic anhydrase,2 ccmm, form 1
|
| 46 | c3fsyC_ |
|
not modelled |
98.9 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:tetrahydrodipicolinate n-succinyltransferase;
PDBTitle: structure of tetrahydrodipicolinate n-succinyltransferase2 (rv1201c;dapd) in complex with succinyl-coa from mycobacterium3 tuberculosis
|
| 47 | c3r1wA_ |
|
not modelled |
98.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of a carbonic anhydrase from a crude oil degrading2 psychrophilic library
|
| 48 | c3d98A_ |
|
not modelled |
98.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
| 49 | c2qkxA_ |
|
not modelled |
98.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
| 50 | c1yp3C_ |
|
not modelled |
98.6 |
10 |
PDB header:transferase
Chain: C: PDB Molecule:glucose-1-phosphate adenylyltransferase small
PDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
|
| 51 | d1yp2a1 |
|
not modelled |
98.5 |
11 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 52 | c2rijA_ |
|
not modelled |
98.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter3 jejuni at 1.90 a resolution
|
| 53 | d1fxja1 |
|
not modelled |
98.3 |
14 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 54 | c1fwyA_ |
|
not modelled |
98.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
|
| 55 | c3brkX_ |
|
not modelled |
98.0 |
19 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from2 agrobacterium tumefaciens
|
| 56 | d2jnga1 |
|
not modelled |
94.5 |
15 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:CPH domain |
| 57 | c2jufA_ |
|
not modelled |
94.4 |
15 |
PDB header:gene regulation
Chain: A: PDB Molecule:p53-associated parkin-like cytoplasmic protein;
PDBTitle: nmr solution structure of parc cph domain. nesg target2 hr3443b/sgc-toronto
|
| 58 | c2zv4O_ |
|
not modelled |
42.8 |
21 |
PDB header:structural protein
Chain: O: PDB Molecule:major vault protein;
PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
|
| 59 | c3anwB_ |
|
not modelled |
41.3 |
19 |
PDB header:replication
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: a protein complex essential initiation of dna replication
|
| 60 | c3gf5A_ |
|
not modelled |
41.0 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:major vault protein;
PDBTitle: crystal structure of the p21 r1-r7 n-terminal domain of murine mvp
|
| 61 | c1y7xA_ |
|
not modelled |
40.8 |
29 |
PDB header:structural protein, protein binding
Chain: A: PDB Molecule:major vault protein;
PDBTitle: solution structure of a two-repeat fragment of major vault2 protein
|
| 62 | c2i5kB_ |
|
not modelled |
23.3 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of ugp1p
|
| 63 | c1vi7A_ |
|
not modelled |
21.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yigz;
PDBTitle: crystal structure of an hypothetical protein
|
| 64 | d2icya1 |
|
not modelled |
15.0 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 65 | c1whsB_ |
|
not modelled |
14.7 |
27 |
PDB header:serine carboxypeptidase
Chain: B: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: structure of the complex of l-benzylsuccinate with wheat serine2 carboxypeptidase ii at 2.0 angstroms resolution
|
| 66 | c2k6lA_ |
|
not modelled |
13.4 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
|
| 67 | d1i1ja_ |
|
not modelled |
10.8 |
13 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 68 | d1yw6a1 |
|
not modelled |
10.8 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 69 | d1bb9a_ |
|
not modelled |
10.8 |
17 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 70 | c1bb9A_ |
|
not modelled |
10.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:amphiphysin 2;
PDBTitle: crystal structure of the sh3 domain from rat amphiphysin 2
|
| 71 | d1loua_ |
|
not modelled |
10.7 |
43 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 72 | d2qalf1 |
|
not modelled |
10.3 |
43 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 73 | d1qjha_ |
|
not modelled |
10.1 |
43 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 74 | d2g9da1 |
|
not modelled |
9.9 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 75 | c2jkuA_ |
|
not modelled |
9.8 |
38 |
PDB header:ligase
Chain: A: PDB Molecule:propionyl-coa carboxylase alpha chain,
PDBTitle: crystal structure of the n-terminal region of the biotin2 acceptor domain of human propionyl-coa carboxylase
|
| 76 | d1jw3a_ |
|
not modelled |
9.8 |
10 |
Fold:MTH1598-like
Superfamily:MTH1598-like
Family:MTH1598-like |
| 77 | d1qlya_ |
|
not modelled |
9.6 |
22 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 78 | c2ow7A_ |
|
not modelled |
9.2 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase 2;
PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
|
| 79 | c1scfA_ |
|
not modelled |
8.9 |
33 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:stem cell factor;
PDBTitle: human recombinant stem cell factor
|
| 80 | d1scfa_ |
|
not modelled |
8.9 |
33 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Short-chain cytokines |
| 81 | d1wb8a1 |
|
not modelled |
8.8 |
38 |
Fold:Long alpha-hairpin
Superfamily:Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family:Fe,Mn superoxide dismutase (SOD), N-terminal domain |
| 82 | c2qbbF_ |
|
not modelled |
8.8 |
43 |
PDB header:ribosome
Chain: F: PDB Molecule:30s ribosomal protein s6;
PDBTitle: crystal structure of the bacterial ribosome from2 escherichia coli in complex with gentamicin. this file3 contains the 30s subunit of the second 70s ribosome, with4 gentamicin bound. the entire crystal structure contains5 two 70s ribosomes and is described in remark 400.
|
| 83 | d1b06a1 |
|
not modelled |
8.8 |
13 |
Fold:Long alpha-hairpin
Superfamily:Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family:Fe,Mn superoxide dismutase (SOD), N-terminal domain |
| 84 | c1o7dA_ |
|
not modelled |
8.4 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysosomal alpha-mannosidase;
PDBTitle: the structure of the bovine lysosomal a-mannosidase2 suggests a novel mechanism for low ph activation
|
| 85 | d1edya_ |
|
not modelled |
8.4 |
3 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Alpha-macroglobulin receptor domain
Family:Alpha-macroglobulin receptor domain |
| 86 | c1ov3A_ |
|
not modelled |
8.0 |
20 |
PDB header:oxidoreductase activator
Chain: A: PDB Molecule:neutrophil cytosol factor 1;
PDBTitle: structure of the p22phox-p47phox complex
|
| 87 | c3bbnF_ |
|
not modelled |
7.5 |
29 |
PDB header:ribosome
Chain: F: PDB Molecule:ribosomal protein s6;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 88 | d2j5aa1 |
|
not modelled |
7.4 |
29 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 89 | d1ov3a1 |
|
not modelled |
7.3 |
19 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 90 | c1gxsB_ |
|
not modelled |
7.3 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain b;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 91 | d1kk8a1 |
|
not modelled |
7.1 |
50 |
Fold:SH3-like barrel
Superfamily:Myosin S1 fragment, N-terminal domain
Family:Myosin S1 fragment, N-terminal domain |
| 92 | d2fcta1 |
|
not modelled |
6.9 |
32 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 93 | c3iuwA_ |
|
not modelled |
6.8 |
10 |
PDB header:rna binding protein
Chain: A: PDB Molecule:activating signal cointegrator;
PDBTitle: crystal structure of activating signal cointegrator (np_814290.1) from2 enterococcus faecalis v583 at 1.58 a resolution
|
| 94 | d1scfc_ |
|
not modelled |
6.7 |
33 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Short-chain cytokines |
| 95 | c3mp2A_ |
|
not modelled |
6.6 |
67 |
PDB header:hydrolase
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of transmissible gastroenteritis virus papain-like2 protease 1
|
| 96 | c2fe8B_ |
|
not modelled |
6.5 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: sars coronavirus papain-like protease: structure of a viral2 deubiquitinating enzyme
|
| 97 | c2csiA_ |
|
not modelled |
6.4 |
24 |
PDB header:endocytosis/exocytosis
Chain: A: PDB Molecule:rim binding protein 2;
PDBTitle: solution structure of the third sh3 domain of human rim-2 binding protein 2
|
| 98 | c1t0jA_ |
|
not modelled |
6.4 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:voltage-gated calcium channel subunit beta2a;
PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
|
| 99 | d1bv8a_ |
|
not modelled |
6.4 |
9 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Alpha-macroglobulin receptor domain
Family:Alpha-macroglobulin receptor domain |