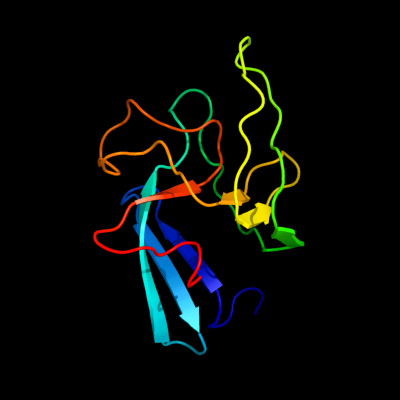
| 1 | c2hklB_
|
|
 |
99.6 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:l,d-transpeptidase;
PDBTitle: crystal structure of enterococcus faecium l,d-2 transpeptidase c442s mutant
|
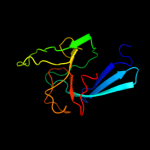
| 2 | d1zata1
|
|
 |
99.5 |
24 |
Fold:L,D-transpeptidase catalytic domain-like
Superfamily:L,D-transpeptidase catalytic domain-like
Family:L,D-transpeptidase catalytic domain-like |
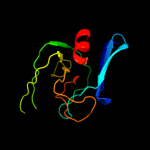
| 3 | d1lbua1
|
|
 |
99.2 |
34 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:Peptidoglycan binding domain, PGBD |
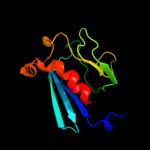
| 4 | c1y7mB_
|
|
 |
99.2 |
29 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein bsu14040;
PDBTitle: crystal structure of the b. subtilis ykud protein at 2 a2 resolution
|
| 5 | c1lbuA_
|
|
 |
99.1 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:muramoyl-pentapeptide carboxypeptidase;
PDBTitle: hydrolase metallo (zn) dd-peptidase
|
| 6 | c3bkhA_
|
|
 |
99.1 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:lytic transglycosylase;
PDBTitle: crystal structure of the bacteriophage phikz lytic2 transglycosylase, gp144
|
| 7 | d1y7ma1
|
|
 |
99.1 |
30 |
Fold:L,D-transpeptidase catalytic domain-like
Superfamily:L,D-transpeptidase catalytic domain-like
Family:L,D-transpeptidase catalytic domain-like |
| 8 | c2bh7A_
|
|
 |
98.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
|
| 9 | c1l6jA_
|
|
 |
97.3 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:matrix metalloproteinase-9;
PDBTitle: crystal structure of human matrix metalloproteinase mmp92 (gelatinase b).
|
| 10 | d1eaka1
|
|
 |
97.1 |
16 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
| 11 | d2ikba1
|
|
 |
97.0 |
28 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:NMB1012-like |
| 12 | d1slma1
|
|
 |
97.0 |
18 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
| 13 | c1slmA_
|
|
 |
97.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:stromelysin-1;
PDBTitle: crystal structure of fibroblast stromelysin-1: the c-truncated human2 proenzyme
|
| 14 | d1su3a1
|
|
 |
96.8 |
22 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
| 15 | c1su3A_
|
|
 |
96.7 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:interstitial collagenase;
PDBTitle: x-ray structure of human prommp-1: new insights into2 collagenase action
|
| 16 | c1eakA_
|
|
 |
96.5 |
22 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:72 kda type iv collagenase;
PDBTitle: catalytic domain of prommp-2 e404q mutant
|
| 17 | d2nr7a1
|
|
 |
96.3 |
21 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:NMB1012-like |
| 18 | d1l6ja1
|
|
 |
96.2 |
23 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
| 19 | c1gxdA_
|
|
 |
93.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:72 kda type iv collagenase;
PDBTitle: prommp-2/timp-2 complex
|
| 20 | c3a05A_
|
|
 |
74.1 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from2 hyperthermophilic archaeon, aeropyrum pernix k1 complex3 with tryptophan
|
| 21 | d2bgxa1 |
|
not modelled |
67.0 |
20 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:Peptidoglycan binding domain, PGBD |
| 22 | d2oo3a1 |
|
not modelled |
49.4 |
27 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:LPG1296-like |
| 23 | c2rjbD_ |
|
not modelled |
45.2 |
23 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein ydcj (sf1787)2 from shigella flexneri which includes domain duf1338.3 northeast structural genomics consortium target sfr276
|
| 24 | c2juoA_ |
|
not modelled |
40.4 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:ga-binding protein alpha chain;
PDBTitle: gabpa ost domain
|
| 25 | c3n9iA_ |
|
not modelled |
38.2 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from yersinia pestis2 co92
|
| 26 | c1wsuA_ |
|
not modelled |
35.6 |
12 |
PDB header:translation/rna
Chain: A: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
|
| 27 | d2ns0a1 |
|
not modelled |
34.4 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:RHA1 ro06458-like |
| 28 | d1r1ga_ |
|
not modelled |
30.2 |
67 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Short-chain scorpion toxins |
| 29 | c1r1gB_ |
|
not modelled |
30.2 |
67 |
PDB header:toxin
Chain: B: PDB Molecule:neurotoxin bmk37;
PDBTitle: crystal structure of the scorpion toxin bmbkttx1
|
| 30 | c1r1gA_ |
|
not modelled |
30.2 |
67 |
PDB header:toxin
Chain: A: PDB Molecule:neurotoxin bmk37;
PDBTitle: crystal structure of the scorpion toxin bmbkttx1
|
| 31 | d2d69a1 |
|
not modelled |
29.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 32 | c2d69B_ |
|
not modelled |
25.9 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
| 33 | c2r5kE_ |
|
not modelled |
25.8 |
27 |
PDB header:viral protein
Chain: E: PDB Molecule:major capsid protein l1;
PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 11
|
| 34 | c3phuA_ |
|
not modelled |
24.5 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:rna-directed rna polymerase l;
PDBTitle: otu domain of crimean congo hemorrhagic fever virus
|
| 35 | c2r5iL_ |
|
not modelled |
23.8 |
24 |
PDB header:viral protein
Chain: L: PDB Molecule:l1 protein;
PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 18
|
| 36 | c3pt2A_ |
|
not modelled |
23.4 |
30 |
PDB header:hydrolase/protein binding
Chain: A: PDB Molecule:rna polymerase;
PDBTitle: structure of a viral otu domain protease bound to ubiquitin
|
| 37 | c1dzlA_ |
|
not modelled |
23.1 |
27 |
PDB header:virus
Chain: A: PDB Molecule:late major capsid protein l1;
PDBTitle: l1 protein of human papillomavirus 16
|
| 38 | d1dzla_ |
|
not modelled |
23.1 |
27 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Group I dsDNA viruses
Family:Papovaviridae-like VP |
| 39 | c3qm2A_ |
|
not modelled |
22.7 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
|
| 40 | c3jxeB_ |
|
not modelled |
22.1 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of pyrococcus horikoshii tryptophanyl-trna2 synthetase in complex with trpamp
|
| 41 | c3hv0A_ |
|
not modelled |
21.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from cryptosporidium parvum
|
| 42 | c2yy5C_ |
|
not modelled |
21.4 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from mycoplasma2 pneumoniae
|
| 43 | d1i6la_ |
|
not modelled |
20.8 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 44 | c2lafA_ |
|
not modelled |
20.4 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:lipoprotein 34;
PDBTitle: nmr solution structure of the n-terminal domain of the e. coli2 lipoprotein bamc
|
| 45 | c2zviB_ |
|
not modelled |
20.2 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
|
| 46 | d1ykwa1 |
|
not modelled |
20.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 47 | d2p0la1 |
|
not modelled |
19.5 |
20 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:LplA-like |
| 48 | c3iyjE_ |
|
not modelled |
19.2 |
18 |
PDB header:virus
Chain: E: PDB Molecule:major capsid protein l1;
PDBTitle: bovine papillomavirus type 1 outer capsid
|
| 49 | d1ej7l1 |
|
not modelled |
18.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 50 | c2qygC_ |
|
not modelled |
17.8 |
16 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
| 51 | c2j5bA_ |
|
not modelled |
17.6 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:tyrosyl-trna synthetase;
PDBTitle: structure of the tyrosyl trna synthetase from acanthamoeba2 polyphaga mimivirus complexed with tyrosynol
|
| 52 | c2ip1A_ |
|
not modelled |
17.2 |
34 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure analysis of s. cerevisiae tryptophanyl trna2 synthetase
|
| 53 | c3elyA_ |
|
not modelled |
17.1 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: wesselsbron virus methyltransferase in complex with adohcy
|
| 54 | c1gehE_ |
|
not modelled |
16.7 |
12 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
| 55 | d3ci0j1 |
|
not modelled |
16.5 |
19 |
Fold:Pili subunits
Superfamily:Pili subunits
Family:EpsJ-like |
| 56 | c2xvsA_ |
|
not modelled |
16.4 |
21 |
PDB header:antitumor protein
Chain: A: PDB Molecule:tetratricopeptide repeat protein 5;
PDBTitle: crystal structure of human ttc5 (strap) c-terminal ob2 domain
|
| 57 | c1telA_ |
|
not modelled |
16.4 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 58 | c2oxtC_ |
|
not modelled |
16.2 |
28 |
PDB header:viral protein
Chain: C: PDB Molecule:nucleoside-2'-o-methyltransferase;
PDBTitle: crystal structure of meaban virus nucleoside-2'-o-2 methyltransferase
|
| 59 | d1geha1 |
|
not modelled |
16.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 60 | d1wuua1 |
|
not modelled |
16.0 |
26 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 61 | c2plyB_ |
|
not modelled |
15.8 |
12 |
PDB header:translation/rna
Chain: B: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: structure of the mrna binding fragment of elongation factor2 selb in complex with secis rna.
|
| 62 | d1rbla1 |
|
not modelled |
15.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 63 | d1pgsa2 |
|
not modelled |
15.4 |
26 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:PHM/PNGase F
Family:Glycosyl-asparaginase |
| 64 | d1iufa2 |
|
not modelled |
14.4 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Centromere-binding |
| 65 | c2i6oA_ |
|
not modelled |
14.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfolobus solfataricus protein tyrosine
PDBTitle: crystal structure of the complex of the archaeal sulfolobus2 ptp-fold phosphatase with phosphopeptides n-g-(p)y-k-n
|
| 66 | d1vlra2 |
|
not modelled |
13.5 |
24 |
Fold:mRNA decapping enzyme DcpS N-terminal domain
Superfamily:mRNA decapping enzyme DcpS N-terminal domain
Family:mRNA decapping enzyme DcpS N-terminal domain |
| 67 | c1zswA_ |
|
not modelled |
13.2 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:glyoxalase family protein;
PDBTitle: crystal structure of bacillus cereus metallo protein from glyoxalase2 family
|
| 68 | c3nwrA_ |
|
not modelled |
13.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 69 | c3nk4B_ |
|
not modelled |
12.8 |
25 |
PDB header:cell adhesion
Chain: B: PDB Molecule:zona pellucida 3;
PDBTitle: crystal structure of full-length sperm receptor zp3 at 2.0 a2 resolution
|
| 70 | c2quiB_ |
|
not modelled |
12.3 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structures of human tryptophanyl-trna synthetase in2 complex with tryptophanamide and atp
|
| 71 | c3hzrD_ |
|
not modelled |
12.1 |
26 |
PDB header:ligase
Chain: D: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase homolog from entamoeba histolytica
|
| 72 | c1r6uB_ |
|
not modelled |
11.9 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of an active fragment of human tryptophanyl-trna2 synthetase with cytokine activity
|
| 73 | d2p5ia1 |
|
not modelled |
11.8 |
19 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:LplA-like |
| 74 | d2z15a1 |
|
not modelled |
11.7 |
16 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 75 | d8ruca1 |
|
not modelled |
11.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 76 | d3bl9a2 |
|
not modelled |
11.4 |
25 |
Fold:mRNA decapping enzyme DcpS N-terminal domain
Superfamily:mRNA decapping enzyme DcpS N-terminal domain
Family:mRNA decapping enzyme DcpS N-terminal domain |
| 77 | c2gezE_ |
|
not modelled |
11.4 |
11 |
PDB header:hydrolase
Chain: E: PDB Molecule:l-asparaginase alpha subunit;
PDBTitle: crystal structure of potassium-independent plant asparaginase
|
| 78 | c3sz3A_ |
|
not modelled |
11.3 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from vibrio cholerae2 with an endogenous tryptophan
|
| 79 | d3e9va1 |
|
not modelled |
11.1 |
23 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 80 | c3focB_ |
|
not modelled |
11.1 |
26 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from giardia lamblia
|
| 81 | c1rcxH_ |
|
not modelled |
11.0 |
17 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 82 | d1atga_ |
|
not modelled |
10.7 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 83 | c1vqzA_ |
|
not modelled |
10.7 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:lipoate-protein ligase, putative;
PDBTitle: crystal structure of a putative lipoate-protein ligase a (sp_1160)2 from streptococcus pneumoniae tigr4 at 1.99 a resolution
|
| 84 | d1bwva1 |
|
not modelled |
10.5 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 85 | c2yfnA_ |
|
not modelled |
10.4 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase-sucrose kinase agask;
PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
|
| 86 | c1bqfA_ |
|
not modelled |
10.0 |
57 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:protein (growth-blocking peptide);
PDBTitle: growth-blocking peptide (gbp) from pseudaletia separata
|
| 87 | c2kimA_ |
|
not modelled |
9.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:o6-methylguanine-dna methyltransferase;
PDBTitle: 1.7-mm microcryoprobe solution nmr structure of an o6-2 methylguanine dna methyltransferase family protein from3 vibrio parahaemolyticus. northeast structural genomics4 consortium target vpr247.
|
| 88 | c3qfwB_ |
|
not modelled |
9.6 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
| 89 | c3prhB_ |
|
not modelled |
8.5 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase val144pro mutant from b. subtilis
|
| 90 | c1iufA_ |
|
not modelled |
8.5 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:centromere abp1 protein;
PDBTitle: low resolution solution structure of the two dna-binding2 domains in schizosaccharomyces pombe abp1 protein
|
| 91 | d1vqza2 |
|
not modelled |
8.4 |
15 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:LplA-like |
| 92 | d1vlya2 |
|
not modelled |
8.1 |
11 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
| 93 | c3tgoD_ |
|
not modelled |
7.9 |
13 |
PDB header:membrane protein
Chain: D: PDB Molecule:lipoprotein 34;
PDBTitle: crystal structure of the e. coli bamcd complex
|
| 94 | c3e4wB_ |
|
not modelled |
7.9 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a 33kda catalase-related protein from2 mycobacterium avium subsp. paratuberculosis. p2(1)2(1)2(1) crystal3 form.
|
| 95 | c1sghA_ |
|
not modelled |
7.8 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:moesin;
PDBTitle: moesin ferm domain bound to ebp50 c-terminal peptide
|
| 96 | c3m5wB_ |
|
not modelled |
7.8 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from2 campylobacter jejuni
|
| 97 | d2ibaa2 |
|
not modelled |
7.7 |
8 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:Urate oxidase (uricase) |
| 98 | d1dgsa1 |
|
not modelled |
7.5 |
17 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:NAD+-dependent DNA ligase, domain 3 |
| 99 | d1r8se_ |
|
not modelled |
7.3 |
22 |
Fold:alpha-alpha superhelix
Superfamily:Sec7 domain
Family:Sec7 domain |