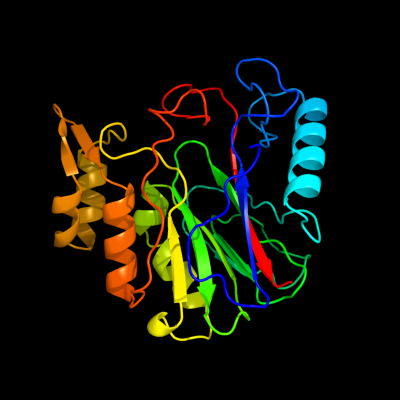
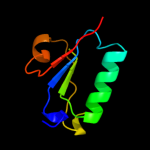
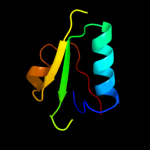
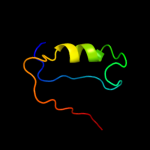
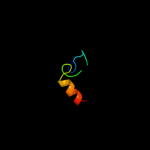
1 d1rw0a_
100.0
87
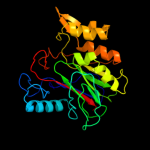
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like2 d1xafa_
100.0
97
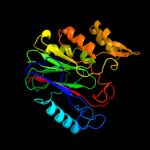
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like3 d1rv9a_
100.0
50
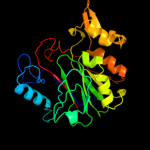
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like4 d1t8ha_
100.0
36
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like5 d1xfja_
100.0
38
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like6 d2f9zc1
93.2
17
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: CheD-like7 d1np7a2
49.0
12
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain8 c3sdsA_
46.9
19
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase, mitochondrial;PDBTitle: crystal structure of a mitochondrial ornithine carbamoyltransferase2 from coccidioides immitis
9 c3giwA_
38.1
9
PDB header: unknown functionChain: A: PDB Molecule: protein of unknown function duf574;PDBTitle: crystal structure of a duf574 family protein (sav_2177) from2 streptomyces avermitilis ma-4680 at 1.45 a resolution
10 c3grfA_
35.2
21
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: x-ray structure of ornithine transcarbamoylase from giardia2 lamblia
11 c3g7kD_
33.4
25
PDB header: isomeraseChain: D: PDB Molecule: 3-methylitaconate isomerase;PDBTitle: crystal structure of methylitaconate-delta-isomerase
12 c2qe6B_
28.1
13
PDB header: transferaseChain: B: PDB Molecule: uncharacterized protein tfu_2867;PDBTitle: crystal structure of a putative methyltransferase (tfu_2867) from2 thermobifida fusca yx at 1.95 a resolution
13 c1kjkA_
26.8
33
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: solution structure of the lambda integrase amino-terminal2 domain
14 d1z1ba1
24.4
33
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: lambda integrase N-terminal domain15 d1owla2
23.8
12
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain16 c1ecjB_
22.4
11
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
17 c2otcA_
21.0
20
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: ornithine transcarbamoylase complexed with n-2 (phosphonacetyl)-l-ornithine
18 d1duvg2
20.3
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase19 c3bd0D_
19.4
21
PDB header: peptide binding proteinChain: D: PDB Molecule: protein memo1;PDBTitle: crystal structure of memo, form ii
20 c2jblC_
17.3
26
PDB header: electron transportChain: C: PDB Molecule: photosynthetic reaction center cytochrome cPDBTitle: photosynthetic reaction center from blastochloris viridis
21 d2i5nc1
not modelled
17.3
26
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Photosynthetic reaction centre (cytochrome subunit)22 d2bo9b1
not modelled
16.1
14
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Latexin-like23 c2w37A_
not modelled
16.1
18
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase, catabolic;PDBTitle: crystal structure of the hexameric catabolic ornithine2 transcarbamylase from lactobacillus hilgardii
24 d1otfa_
not modelled
15.3
15
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like25 c1zrtD_
not modelled
14.4
19
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
26 c2fynH_
not modelled
14.3
19
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
27 c1a1sA_
not modelled
13.6
17
PDB header: transcarbamylaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: ornithine carbamoyltransferase from pyrococcus furiosus
28 d2nvma1
not modelled
13.6
21
Fold: XisI-likeSuperfamily: XisI-likeFamily: XisI-like29 d1dxha2
not modelled
13.4
14
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase30 c2pw0A_
not modelled
12.9
21
PDB header: unknown functionChain: A: PDB Molecule: prpf methylaconitate isomerase;PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
31 c3lyvF_
not modelled
12.4
63
PDB header: chaperoneChain: F: PDB Molecule: ribosome-associated factor y;PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
32 d1vlva2
not modelled
12.1
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase33 d2nlva1
not modelled
11.9
21
Fold: XisI-likeSuperfamily: XisI-likeFamily: XisI-like34 c2qlcC_
not modelled
11.8
15
PDB header: dna binding proteinChain: C: PDB Molecule: dna repair protein radc homolog;PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
35 d1xbta2
not modelled
11.5
21
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger36 d2b8ta2
not modelled
11.3
25
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger37 d2h9fa1
not modelled
11.1
24
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PA0793-like38 c1eysC_
not modelled
10.5
22
PDB header: electron transportChain: C: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
39 d1eysc_
not modelled
10.5
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Photosynthetic reaction centre (cytochrome subunit)40 c2hfqA_
not modelled
10.1
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: nmr structure of protein ne1680 from nitrosomonas europaea:2 northeast structural genomics consortium target net5
41 d2hfqa1
not modelled
10.1
21
Fold: NE1680-likeSuperfamily: NE1680-likeFamily: NE1680-like42 c1p84D_
not modelled
9.8
13
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
43 d1wjpa2
not modelled
9.8
45
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H244 d1xkia_
not modelled
9.8
16
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like45 d1otha2
not modelled
9.6
13
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase46 c3d7qB_
not modelled
9.5
11
PDB header: unknown functionChain: B: PDB Molecule: xisi protein-like;PDBTitle: crystal structure of a xisi-like protein (npun_ar114) from nostoc2 punctiforme pcc 73102 at 2.30 a resolution
47 d1bjpa_
not modelled
9.5
12
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like48 c2f9iC_
not modelled
9.1
13
PDB header: transferaseChain: C: PDB Molecule: acetyl-coenzyme a carboxylase carboxylPDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
49 d2hrva_
not modelled
9.1
28
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold50 c3by0B_
not modelled
8.8
18
PDB header: ligand binding proteinChain: B: PDB Molecule: neutrophil gelatinase-associated lipocalin;PDBTitle: crystal structure of siderocalin (ngal, lipocalin 2) w79a-r81a2 complexed with ferric enterobactin
51 c1z8rA_
not modelled
8.6
20
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
52 d2ch5a2
not modelled
8.6
18
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like53 c3rlhA_
not modelled
8.5
25
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of a class ii phospholipase d from loxosceles2 intermedia venom
54 c3updA_
not modelled
8.5
15
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: 2.9 angstrom crystal structure of ornithine carbamoyltransferase2 (argf) from vibrio vulnificus
55 c3ot4F_
not modelled
8.4
23
PDB header: hydrolaseChain: F: PDB Molecule: putative isochorismatase;PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
56 c3tvsA_
not modelled
8.3
10
PDB header: signaling proteinChain: A: PDB Molecule: cryptochrome-1;PDBTitle: structure of full-length drosophila cryptochrome
57 d1b0aa2
not modelled
8.1
16
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase58 d2nwva1
not modelled
8.0
13
Fold: XisI-likeSuperfamily: XisI-likeFamily: XisI-like59 c3gbcA_
not modelled
7.9
18
PDB header: hydrolaseChain: A: PDB Molecule: pyrazinamidase/nicotinamidas pnca;PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
60 c2op8A_
not modelled
7.9
23
PDB header: isomeraseChain: A: PDB Molecule: probable tautomerase ywhb;PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
61 c3m20A_
not modelled
7.6
8
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
62 d1ppjd1
not modelled
7.6
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain63 d3cx5d1
not modelled
7.6
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain64 c3ecyA_
not modelled
7.5
8
PDB header: hydrolaseChain: A: PDB Molecule: cg4584-pa, isoform a (bcdna.ld08534);PDBTitle: crystal structural analysis of drosophila melanogaster dutpase
65 c1gph1_
not modelled
7.3
11
PDB header: transferase(glutamine amidotransferase)Chain: 1: PDB Molecule: glutamine phosphoribosyl-pyrophosphate amidotransferase;PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
66 d1ml4a2
not modelled
7.0
16
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase67 d1mwwa_
not modelled
6.7
8
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: Hypothetical protein HI1388.168 c3ry0A_
not modelled
6.6
8
PDB header: isomeraseChain: A: PDB Molecule: putative tautomerase;PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
69 c3cwbQ_
not modelled
6.5
13
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
70 d2f9ya1
not modelled
6.4
11
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain71 c2rgwD_
not modelled
6.3
14
PDB header: transferaseChain: D: PDB Molecule: aspartate carbamoyltransferase;PDBTitle: catalytic subunit of m. jannaschii aspartate2 transcarbamoylase
72 c2l5pA_
not modelled
6.2
11
PDB header: transport proteinChain: A: PDB Molecule: lipocalin 12;PDBTitle: solution nmr structure of protein lipocalin 12 from rat epididymis
73 c3gd5D_
not modelled
6.2
22
PDB header: transferaseChain: D: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
74 c3uzuA_
not modelled
6.2
19
PDB header: transferaseChain: A: PDB Molecule: ribosomal rna small subunit methyltransferase a;PDBTitle: the structure of the ribosomal rna small subunit methyltransferase a2 from burkholderia pseudomallei
75 c2f9rC_
not modelled
6.1
25
PDB header: hydrolaseChain: C: PDB Molecule: sphingomyelinase d 1;PDBTitle: crystal structure of the inactive state of the smase i, a2 sphingomyelinase d from loxosceles laeta venom
76 c2ormA_
not modelled
6.0
23
PDB header: isomeraseChain: A: PDB Molecule: probable tautomerase hp0924;PDBTitle: crystal structure of the 4-oxalocrotonate tautomerase homologue dmpi2 from helicobacter pylori.
77 c2z8nB_
not modelled
5.9
24
PDB header: lyaseChain: B: PDB Molecule: 27.5 kda virulence protein;PDBTitle: structural basis for the catalytic mechanism of phosphothreonine lyase
78 c2bo9B_
not modelled
5.9
13
PDB header: hydrolaseChain: B: PDB Molecule: human latexin;PDBTitle: human carboxypeptidase a4 in complex with human latexin.
79 c3mb2G_
not modelled
5.9
8
PDB header: isomeraseChain: G: PDB Molecule: 4-oxalocrotonate tautomerase family enzyme - alpha subunit;PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
80 c2x4kB_
not modelled
5.8
15
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
81 c2orvB_
not modelled
5.8
16
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: human thymidine kinase 1 in complex with tp4a
82 d1x71a1
not modelled
5.7
18
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like83 c3nglA_
not modelled
5.7
16
PDB header: oxidoreductase, hydrolaseChain: A: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
84 c1hynQ_
not modelled
5.7
15
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
85 c1jmxA_
not modelled
5.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: amine dehydrogenase;PDBTitle: crystal structure of a quinohemoprotein amine dehydrogenase2 from pseudomonas putida
86 c3hcyB_
not modelled
5.7
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative two-component sensor histidine kinase protein;PDBTitle: the crystal structure of the domain of putative two-component sensor2 histidine kinase protein from sinorhizobium meliloti 1021
87 d1gph11
not modelled
5.7
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)88 c3uinD_
not modelled
5.6
50
PDB header: ligase/isomerase/protein bindingChain: D: PDB Molecule: e3 sumo-protein ligase ranbp2;PDBTitle: complex between human rangap1-sumo2, ubc9 and the ir1 domain from2 ranbp2
89 c3abfB_
not modelled
5.5
19
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of a 4-oxalocrotonate tautomerase homologue2 (tthb242)
90 d1pvva2
not modelled
5.4
20
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase91 c3bmaC_
not modelled
5.4
57
PDB header: ligaseChain: C: PDB Molecule: d-alanyl-lipoteichoic acid synthetase;PDBTitle: crystal structure of d-alanyl-lipoteichoic acid synthetase from2 streptococcus pneumoniae r6
92 d1yupa1
not modelled
5.3
9
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like93 c2h0rD_
not modelled
5.3
5
PDB header: hydrolaseChain: D: PDB Molecule: nicotinamidase;PDBTitle: structure of the yeast nicotinamidase pnc1p
94 c2kswA_
not modelled
5.2
33
PDB header: hydrolase inhibitorChain: A: PDB Molecule: oryctin;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for oryctin
95 c3on9B_
not modelled
5.2
22
PDB header: viral proteinChain: B: PDB Molecule: tumour necrosis factor receptor;PDBTitle: the secret domain from ectromelia virus
96 c3e2iA_
not modelled
5.1
11
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from s. aureus
97 d1ecfa1
not modelled
5.1
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)98 d1hynp_
not modelled
5.0
15
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain