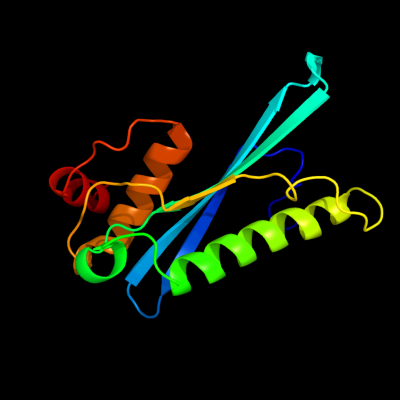
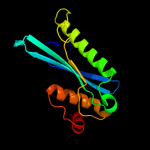
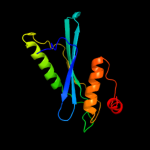
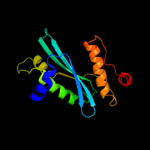
1 d1euva_
97.9
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like2 d2g4da1
97.6
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like3 d2bkra1
97.5
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like4 c2oixA_
97.3
15
PDB header: hydrolaseChain: A: PDB Molecule: xanthomonas outer protein d;PDBTitle: xanthomonas xopd c470a mutant
5 c3eayA_
97.2
13
PDB header: hydrolaseChain: A: PDB Molecule: sentrin-specific protease 7;PDBTitle: crystal structure of the human senp7 catalytic domain
6 d1th0a_
97.0
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like7 d2iy1a1
96.8
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like8 d1nlna_
68.5
26
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like9 d1xd3a_
58.4
16
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Ubiquitin carboxyl-terminal hydrolase UCH-L10 d2cpqa1
56.6
38
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)11 d2etla1
55.7
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Ubiquitin carboxyl-terminal hydrolase UCH-L12 c2wdtA_
46.9
22
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase l3;PDBTitle: crystal structure of plasmodium falciparum uchl3 in complex2 with the suicide inhibitor ubvme
13 c3a7sA_
24.6
12
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase isozyme l5;PDBTitle: catalytic domain of uch37
14 c3mstA_
17.5
23
PDB header: transport proteinChain: A: PDB Molecule: putative nitrate transport protein;PDBTitle: crystal structure of a putative nitrate transport protein (tvn0104)2 from thermoplasma volcanium at 1.35 a resolution
15 c3ihrA_
15.9
14
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase isozyme l5;PDBTitle: crystal structure of uch37
16 d1cjaa_
15.0
15
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Actin-fragmin kinase, catalytic domain17 c3zrhA_
13.8
23
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin thioesterase zranb1;PDBTitle: crystal structure of the lys29, lys33-linkage-specific trabid otu2 deubiquitinase domain reveals an ankyrin-repeat ubiquitin binding3 domain (ankubd)
18 c3dcyA_
13.4
17
PDB header: apoptosis regulatorChain: A: PDB Molecule: regulator protein;PDBTitle: crystal structure a tp53-induced glycolysis and apoptosis2 regulator protein from homo sapiens.
19 c2qqpD_
13.0
27
PDB header: virusChain: D: PDB Molecule: small capsid protein;PDBTitle: crystal structure of authentic providence virus
20 c2of6C_
10.7
50
PDB header: virusChain: C: PDB Molecule: envelope glycoprotein e;PDBTitle: structure of immature west nile virus
21 d1s2xa_
not modelled
10.7
47
Fold: STAT-likeSuperfamily: Cag-ZFamily: Cag-Z22 c1s2xA_
not modelled
10.7
47
PDB header: unknown functionChain: A: PDB Molecule: cag-z;PDBTitle: crystal structure of cag-z from helicobacter pylori
23 d1z21a1
not modelled
10.6
21
Fold: Type III secretion system domainSuperfamily: Type III secretion system domainFamily: YopR Core24 c2i3eA_
not modelled
10.3
20
PDB header: hydrolaseChain: A: PDB Molecule: g-rich;PDBTitle: solution structure of catalytic domain of goldfish rich2 protein
25 d1aina_
not modelled
10.0
23
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin26 c1uzgA_
not modelled
9.9
50
PDB header: viral proteinChain: A: PDB Molecule: major envelope protein e;PDBTitle: crystal structure of the dengue type 3 virus envelope2 protein
27 d1u7za_
not modelled
9.9
15
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like28 c2ka2A_
not modelled
9.8
43
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
29 c2ka1B_
not modelled
9.8
43
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
30 d1zrua3
not modelled
9.7
27
Fold: Pseudo beta-prismSuperfamily: Bacteriophage trimeric proteins domainFamily: Lactophage receptor-binding protein N-terminal domain31 c2ka2B_
not modelled
9.3
43
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
32 c2ka1A_
not modelled
9.3
43
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
33 d2arha1
not modelled
9.1
41
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Aq 1966-like34 d1svba2
not modelled
9.0
50
Fold: Viral glycoprotein, central and dimerisation domainsSuperfamily: Viral glycoprotein, central and dimerisation domainsFamily: Viral glycoprotein, central and dimerisation domains35 c3lcnC_
not modelled
8.8
46
PDB header: nuclear proteinChain: C: PDB Molecule: mrna transport factor gfd1;PDBTitle: nab2:gfd1 complex
36 c3lcnD_
not modelled
8.5
46
PDB header: nuclear proteinChain: D: PDB Molecule: mrna transport factor gfd1;PDBTitle: nab2:gfd1 complex
37 d1sjpa2
not modelled
8.3
28
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain38 c2f9jP_
not modelled
8.2
27
PDB header: rna binding proteinChain: P: PDB Molecule: splicing factor 3b subunit 1;PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
39 d1ok8a2
not modelled
8.2
50
Fold: Viral glycoprotein, central and dimerisation domainsSuperfamily: Viral glycoprotein, central and dimerisation domainsFamily: Viral glycoprotein, central and dimerisation domains40 c2j5dA_
not modelled
7.9
50
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
41 c1p58C_
not modelled
7.8
50
PDB header: virusChain: C: PDB Molecule: major envelope protein e;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
42 c3uajA_
not modelled
7.8
50
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
43 d1f74a_
not modelled
7.7
21
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase44 c1urzC_
not modelled
7.7
50
PDB header: virus/viral proteinChain: C: PDB Molecule: envelope protein;PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
45 d1o22a_
not modelled
7.3
70
Fold: Hypothetical protein TM0875Superfamily: Hypothetical protein TM0875Family: Hypothetical protein TM087546 d1ufwa_
not modelled
7.1
23
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD47 d1rmka_
not modelled
6.8
83
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Conotoxin48 c3dxeC_
not modelled
6.7
24
PDB header: protein bindingChain: C: PDB Molecule: amyloid beta a4 protein-binding family b memberPDBTitle: crystal structure of the intracellular domain of human app2 (t668a mutant) in complex with fe65-ptb2
49 c3c6dB_
not modelled
6.6
50
PDB header: virusChain: B: PDB Molecule: polyprotein;PDBTitle: the pseudo-atomic structure of dengue immature virus
50 c3siiA_
not modelled
6.5
35
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: poly(adp-ribose) glycohydrolase;PDBTitle: the x-ray crystal structure of poly(adp-ribose) glycohydrolase bound2 to the inhibitor adp-hpd from thermomonospora curvata
51 c3ogzA_
not modelled
6.1
20
PDB header: transferaseChain: A: PDB Molecule: udp-sugar pyrophosphorylase;PDBTitle: protein structure of usp from l. major in apo-form
52 d1dm5a_
not modelled
6.1
23
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin53 d1srva_
not modelled
5.9
31
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain54 d2ptza1
not modelled
5.8
16
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase55 c1bmxA_
not modelled
5.8
32
PDB header: viral proteinChain: A: PDB Molecule: human immunodeficiency virus type 1 capsid;PDBTitle: hiv-1 capsid protein major homology region peptide analog,2 nmr, 8 structures
56 d1vlya1
not modelled
5.7
15
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain57 c2vc6A_
not modelled
5.4
22
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of mosa from s. meliloti with pyruvate bound
58 d1sx5a_
not modelled
5.3
21
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease EcoRV59 c1gp8A_
not modelled
5.2
13
PDB header: viral proteinChain: A: PDB Molecule: protein (scaffolding protein);PDBTitle: nmr solution structure of the coat protein-binding domain2 of bacteriophage p22 scaffolding protein
60 d1t6t1_
not modelled
5.2
8
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain61 d1k8ga2
not modelled
5.2
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB62 c1w9zB_
not modelled
5.1
35
PDB header: virus coat proteinChain: B: PDB Molecule: vp9;PDBTitle: structure of bannavirus vp9