1 c3b9yA_
93.7
15
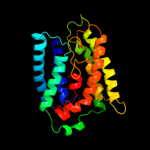
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
2 d1u7ga_
82.0
17
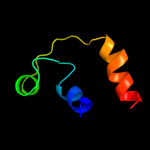
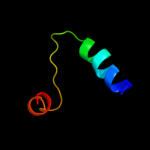
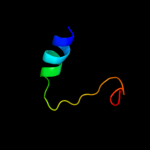
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter3 c3r2uC_
53.6
22
PDB header: hydrolaseChain: C: PDB Molecule: metallo-beta-lactamase family protein;PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
4 d2p97a1
52.1
24
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Ava3068-like5 c2xf4A_
50.9
27
PDB header: hydrolaseChain: A: PDB Molecule: hydroxyacylglutathione hydrolase;PDBTitle: crystal structure of salmonella enterica serovar2 typhimurium ycbl
6 c2gcuD_
47.1
21
PDB header: hydrolaseChain: D: PDB Molecule: putative hydroxyacylglutathione hydrolase 3;PDBTitle: x-ray structure of gene product from arabidopsis thaliana2 at1g53580
7 d1mqoa_
45.7
19
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase8 c3tp9B_
38.7
20
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase and rhodanese domain protein;PDBTitle: crystal structure of alicyclobacillus acidocaldarius protein with2 beta-lactamase and rhodanese domains
9 c3hd6A_
38.4
13
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
10 d1nxca_
36.2
19
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain11 d1xm8a_
35.9
16
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)12 d1qh5a_
34.9
7
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)13 d1znba_
34.8
18
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase14 c2zwrA_
34.2
35
PDB header: hydrolaseChain: A: PDB Molecule: metallo-beta-lactamase superfamily protein;PDBTitle: crystal structure of ttha1623 from thermus thermophilus hb8
15 d1dl2a_
31.1
17
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain16 d2aioa1
30.2
21
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase17 d1bh9b_
28.7
19
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs18 c3hnnD_
27.1
14
PDB header: oxidoreductaseChain: D: PDB Molecule: putative diflavin flavoprotein a 5;PDBTitle: crystal structure of putative diflavin flavoprotein a 5 (fragment 1-2 254) from nostoc sp. pcc 7120, northeast structural genomics3 consortium target nsr435a
19 d2qeda1
25.1
14
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)20 c3bboU_
24.8
14
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein l22;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
21 c2zo4A_
not modelled
24.6
32
PDB header: hydrolaseChain: A: PDB Molecule: metallo-beta-lactamase family protein;PDBTitle: crystal structure of metallo-beta-lactamase family protein ttha14292 from thermus thermophilus hb8
22 c1ze2B_
not modelled
24.2
29
PDB header: lyase/rnaChain: B: PDB Molecule: trna pseudouridine synthase b;PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
23 d1m2xa_
not modelled
23.6
21
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase24 d1bk8a_
not modelled
21.8
57
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Plant defensins25 d2gycq1
not modelled
21.7
36
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L2226 c1s1iN_
not modelled
20.6
33
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l17-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
27 d2ey4a2
not modelled
20.0
30
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB28 c3l6nA_
not modelled
19.6
21
PDB header: hydrolaseChain: A: PDB Molecule: metallo-beta-lactamase;PDBTitle: crystal structure of metallo-beta-lactamase ind-7
29 c1krfA_
not modelled
19.4
19
PDB header: hydrolaseChain: A: PDB Molecule: mannosyl-oligosaccharide alpha-1,2-mannosidase;PDBTitle: structure of p. citrinum alpha 1,2-mannosidase reveals the basis for2 differences in specificity of the er and golgi class i enzymes
30 d2ri9a1
not modelled
19.4
19
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain31 c1vmeB_
not modelled
18.9
17
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
32 d1r3ea2
not modelled
18.4
29
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB33 d1ycga2
not modelled
18.0
18
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: ROO N-terminal domain-like34 d2gmna1
not modelled
17.8
21
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase35 c2yz3B_
not modelled
16.8
21
PDB header: hydrolaseChain: B: PDB Molecule: metallo-beta-lactamase;PDBTitle: crystallographic investigation of inhibition mode of the2 vim-2 metallo-beta-lactamase from pseudomonas aeruginosa3 with mercaptocarboxylate inhibitor
36 d1hcua_
not modelled
16.7
21
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain37 c2b2hA_
not modelled
16.0
10
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter;PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
38 c3pisA_
not modelled
15.8
25
PDB header: hydrolase inhibitorChain: A: PDB Molecule: kazal-type serine protease inhibitor spi-1;PDBTitle: crystal structure of carcinoscorpius rotundicauda serine protease2 inhibitor domain 1
39 c2p18A_
not modelled
15.6
8
PDB header: hydrolaseChain: A: PDB Molecule: glyoxalase ii;PDBTitle: crystal structure of the leishmania infantum glyoxalase ii
40 d1sgva2
not modelled
15.5
26
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB41 d1k8wa5
not modelled
15.5
30
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB42 d2nn6h2
not modelled
15.4
33
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like43 c1g6iA_
not modelled
15.1
16
PDB header: hydrolaseChain: A: PDB Molecule: class i alpha-1,2-mannosidase;PDBTitle: crystal structure of the yeast alpha-1,2-mannosidase with bound 1-2 deoxymannojirimycin at 1.59 a resolution
44 c1k8wA_
not modelled
15.0
30
PDB header: lyase/rnaChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
45 d1ko3a_
not modelled
15.0
23
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase46 c1x9dA_
not modelled
14.8
14
PDB header: hydrolaseChain: A: PDB Molecule: endoplasmic reticulum mannosyl-oligosaccharide 1,PDBTitle: crystal structure of human class i alpha-1,2-mannosidase in2 complex with thio-disaccharide substrate analogue
47 d1x9da1
not modelled
14.8
14
Fold: alpha/alpha toroidSuperfamily: Seven-hairpin glycosidasesFamily: Class I alpha-1;2-mannosidase, catalytic domain48 d1jv1a_
not modelled
14.7
25
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase49 d2foka4
not modelled
14.6
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease FokI, C-terminal (catalytic) domain50 d1pcfa_
not modelled
14.5
42
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: Transcriptional coactivator PC4 C-terminal domain51 d2icya2
not modelled
14.3
40
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase52 d1ayja_
not modelled
14.1
57
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Plant defensins53 c3uaiA_
not modelled
13.9
33
PDB header: isomerase/chaperoneChain: A: PDB Molecule: h/aca ribonucleoprotein complex subunit 4;PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
54 c2fhxB_
not modelled
13.8
28
PDB header: hydrolase, metal binding proteinChain: B: PDB Molecule: spm-1;PDBTitle: pseudomonas aeruginosa spm-1 metallo-beta-lactamase
55 d1f0la3
not modelled
13.4
67
Fold: Toxins' membrane translocation domainsSuperfamily: Diphtheria toxin, middle domainFamily: Diphtheria toxin, middle domain56 c2q9uB_
not modelled
13.3
10
PDB header: oxidoreductaseChain: B: PDB Molecule: a-type flavoprotein;PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
57 c3oc9A_
not modelled
13.3
33
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: crystal structure of putative udp-n-acetylglucosamine2 pyrophosphorylase from entamoeba histolytica
58 d1gu2a_
not modelled
13.1
46
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c59 d1b0aa2
not modelled
12.9
30
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase60 d2apoa2
not modelled
12.5
38
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB61 c3pesA_
not modelled
12.4
67
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein gp49;PDBTitle: crystal structure of uncharacterized protein from pseudomonas phage2 yua
62 c3jywN_
not modelled
11.7
33
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l17(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
63 d1vm8a_
not modelled
11.4
25
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase64 c2e55D_
not modelled
11.1
43
PDB header: transferaseChain: D: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: structure of aq2163 protein from aquifex aeolicus
65 c3lvzA_
not modelled
11.0
17
PDB header: hydrolaseChain: A: PDB Molecule: blr6230 protein;PDBTitle: new refinement of the crystal structure of bjp-1, a subclass b32 metallo-beta-lactamase of bradyrhizobium japonicum
66 c2oefA_
not modelled
11.0
42
PDB header: transferaseChain: A: PDB Molecule: utp-glucose-1-phosphate uridylyltransferase 2,PDBTitle: open and closed structures of the udp-glucose2 pyrophosphorylase from leishmania major
67 c3h0dB_
not modelled
10.8
15
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
68 d1e5da2
not modelled
10.7
18
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: ROO N-terminal domain-like69 c1sgvA_
not modelled
10.6
29
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: structure of trna psi55 pseudouridine synthase (trub)
70 c2ey4A_
not modelled
10.4
33
PDB header: isomerase/biosynthetic proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
71 c4a17Q_
not modelled
10.3
22
PDB header: ribosomeChain: Q: PDB Molecule: rpl17;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
72 d1jjta_
not modelled
10.2
14
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Zn metallo-beta-lactamase73 c3gueB_
not modelled
9.9
42
PDB header: transferaseChain: B: PDB Molecule: utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: crystal structure of udp-glucose phosphorylase from trypanosoma2 brucei, (tb10.389.0330)
74 d1vqor1
not modelled
9.8
11
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L2275 c3fvyA_
not modelled
9.6
36
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl-peptidase 3;PDBTitle: crystal structure of human dipeptidyl peptidase iii
76 c2q4jB_
not modelled
9.3
33
PDB header: transferaseChain: B: PDB Molecule: probable utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
77 d1ecia_
not modelled
9.3
50
Fold: Ectatomin subunitsSuperfamily: Ectatomin subunitsFamily: Ectatomin subunits78 c3sd9B_
not modelled
9.3
19
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase;PDBTitle: crystal structure of serratia fonticola sfh-i: source of the2 nucleophile in the catalytic mechanism of mono-zinc metallo-beta-3 lactamases
79 c2yqsA_
not modelled
9.1
25
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: crystal structure of uridine-diphospho-n-acetylglucosamine2 pyrophosphorylase from candida albicans, in the product-binding form
80 c3dmpD_
not modelled
9.1
29
PDB header: transferaseChain: D: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: 2.6 a crystal structure of uracil phosphoribosyltransferase2 from burkholderia pseudomallei
81 c3spuB_
not modelled
9.0
14
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase ndm-1;PDBTitle: apo ndm-1 crystal structure
82 d1i5ea_
not modelled
8.9
33
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)83 c2i5kB_
not modelled
8.9
33
PDB header: transferaseChain: B: PDB Molecule: utp--glucose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of ugp1p
84 c1ychD_
not modelled
8.7
16
PDB header: oxidoreductaseChain: D: PDB Molecule: nitric oxide reductase;PDBTitle: x-ray crystal structures of moorella thermoacetica fpra.2 novel diiron site structure and mechanistic insights into3 a scavenging nitric oxide reductase
85 d1i4ja_
not modelled
8.6
33
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L2286 c2ftcM_
not modelled
8.5
29
PDB header: ribosomeChain: M: PDB Molecule: mitochondrial ribosomal protein l22 isoform a;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
87 c2apoA_
not modelled
8.4
38
PDB header: isomerase/rna binding proteinChain: A: PDB Molecule: probable trna pseudouridine synthase b;PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
88 c3cskA_
not modelled
8.4
29
PDB header: hydrolaseChain: A: PDB Molecule: probable dipeptidyl-peptidase 3;PDBTitle: structure of dpp iii from saccharomyces cerevisiae
89 d1a4ia2
not modelled
8.4
30
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase90 c3mv2A_
not modelled
8.3
14
PDB header: protein transportChain: A: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of a-cop in complex with e-cop
91 c2fs1A_
not modelled
8.3
33
PDB header: protein bindingChain: A: PDB Molecule: psd-1;PDBTitle: solution structure of psd-1
92 d1gjsa_
not modelled
8.3
33
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: GA module, an albumin-binding domain93 c2zvrA_
not modelled
8.1
36
PDB header: isomeraseChain: A: PDB Molecule: uncharacterized protein tm_0416;PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
94 d2q0ia1
not modelled
8.0
35
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: PqsE-like95 c2zkrr_
not modelled
8.0
22
PDB header: ribosomal protein/rnaChain: R: PDB Molecule: rna expansion segment es39 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
96 c1e5dA_
not modelled
7.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: rubredoxin\:oxygen oxidoreductase;PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe2 desulfovibrio gigas
97 d1edza2
not modelled
7.9
30
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase98 c3adrA_
not modelled
7.7
21
PDB header: signaling proteinChain: A: PDB Molecule: putative uncharacterized protein st1585;PDBTitle: the first crystal structure of an archaeal metallo-beta-lactamase2 superfamily protein; st1585 from sulfolobus tokodaii
99 c3iz5V_
not modelled
7.7
22
PDB header: ribosomeChain: V: PDB Molecule: 60s ribosomal protein l17 (l22p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome