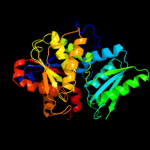
| 1 | d1j0aa_ |
|
 |
100.0 |
40 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
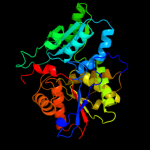
| 2 | d1f2da_ |
|
 |
100.0 |
32 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
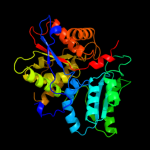
| 3 | d1tyza_ |
|
 |
100.0 |
38 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
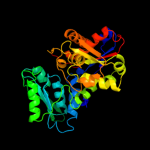
| 4 | c1tdjA_ |
|
 |
100.0 |
22 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 5 | c3l6cA_ |
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:serine racemase;
PDBTitle: x-ray crystal structure of rat serine racemase in complex with2 malonate a potent inhibitor
|
| 6 | c3iauA_ |
|
 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:threonine deaminase;
PDBTitle: the structure of the processed form of threonine deaminase isoform 22 from solanum lycopersicum
|
| 7 | c3pc3A_ |
|
 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:cg1753, isoform a;
PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
|
| 8 | d1v71a1 |
|
 |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 9 | d1z7wa1 |
|
 |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 10 | c2d1fA_ |
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: structure of mycobacterium tuberculosis threonine synthase
|
| 11 | d1jbqa_ |
|
 |
100.0 |
19 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 12 | c1jbqD_ |
|
 |
100.0 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:cystathionine beta-synthase;
PDBTitle: structure of human cystathionine beta-synthase: a unique pyridoxal 5'-2 phosphate dependent hemeprotein
|
| 13 | d1pwha_ |
|
 |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 14 | c2gn0A_ |
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:threonine dehydratase catabolic;
PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
|
| 15 | d1tdja1 |
|
 |
100.0 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 16 | d1wkva1 |
|
 |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 17 | d1p5ja_ |
|
 |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 18 | c1p5jA_ |
|
 |
100.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:l-serine dehydratase;
PDBTitle: crystal structure analysis of human serine dehydratase
|
| 19 | c2pqmA_ |
|
 |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
|
| 20 | d1ve1a1 |
|
 |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 21 | d1v7ca_ |
|
not modelled |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 22 | d1ve5a1 |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 23 | c2zsjB_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
|
| 24 | c3dwgA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase b;
PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
|
| 25 | d2bhsa1 |
|
not modelled |
100.0 |
19 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 26 | c1x1qA_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
|
| 27 | c2rkbE_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:serine dehydratase-like;
PDBTitle: serine dehydratase like-1 from human cancer cells
|
| 28 | c3r0zA_ |
|
not modelled |
100.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:d-serine dehydratase;
PDBTitle: crystal structure of apo d-serine deaminase from salmonella2 typhimurium
|
| 29 | c2q3bA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase a;
PDBTitle: 1.8 a resolution crystal structure of o-acetylserine sulfhydrylase2 (oass) holoenzyme from mycobacterium tuberculosis
|
| 30 | d1qopb_ |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 31 | d1fcja_ |
|
not modelled |
100.0 |
15 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 32 | d1o58a_ |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 33 | d1v8za1 |
|
not modelled |
100.0 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 34 | d1y7la1 |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 35 | c2eguA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of o-acetylserine sulfhydrase from geobacillus2 kaustophilus hta426
|
| 36 | d1e5xa_ |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 37 | c2o2jA_ |
|
not modelled |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain2 dimer (apoform)
|
| 38 | c3v7nA_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase (thrc) from from burkholderia2 thailandensis
|
| 39 | d1vb3a1 |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 40 | d1kl7a_ |
|
not modelled |
99.8 |
15 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 41 | d1hyua1 |
|
not modelled |
79.5 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 42 | d1fl2a1 |
|
not modelled |
76.9 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 43 | d1trba1 |
|
not modelled |
74.7 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 44 | c3ot4F_ |
|
not modelled |
65.9 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 45 | d1vdca1 |
|
not modelled |
65.5 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 46 | d1seza1 |
|
not modelled |
64.2 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 47 | c1djnB_ |
|
not modelled |
61.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 48 | d1j8yf2 |
|
not modelled |
61.1 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 49 | d3lada1 |
|
not modelled |
61.0 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 50 | d2iida1 |
|
not modelled |
60.2 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 51 | c3gmbB_ |
|
not modelled |
59.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic2 acid oxygenase
|
| 52 | d1reoa1 |
|
not modelled |
58.9 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 53 | d1o8ca2 |
|
not modelled |
58.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 54 | d1ebda1 |
|
not modelled |
58.5 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 55 | c2ywlA_ |
|
not modelled |
58.4 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase related protein;
PDBTitle: crystal structure of thioredoxin reductase-related protein ttha03702 from thermus thermophilus hb8
|
| 56 | c2e1mA_ |
|
not modelled |
58.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-glutamate oxidase;
PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
|
| 57 | d1dxla1 |
|
not modelled |
57.9 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 58 | d2i0za1 |
|
not modelled |
57.8 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 59 | c3r2jC_ |
|
not modelled |
57.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
| 60 | c3c7cB_ |
|
not modelled |
56.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
| 61 | d1xpma2 |
|
not modelled |
55.8 |
23 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 62 | c1qzwC_ |
|
not modelled |
54.6 |
14 |
PDB header:signaling protein/rna
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
|
| 63 | c3allA_ |
|
not modelled |
54.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
|
| 64 | d1lvla1 |
|
not modelled |
54.3 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 65 | d1ryia1 |
|
not modelled |
53.0 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 66 | d1wd5a_ |
|
not modelled |
53.0 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 67 | d1ojta1 |
|
not modelled |
52.2 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 68 | c3dm5A_ |
|
not modelled |
52.1 |
17 |
PDB header:rna binding protein, transport protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
|
| 69 | d1pvva2 |
|
not modelled |
51.5 |
16 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 70 | c1j8yF_ |
|
not modelled |
51.3 |
12 |
PDB header:signaling protein
Chain: F: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: signal recognition particle conserved gtpase domain from a.2 ambivalens t112a mutant
|
| 71 | d1t57a_ |
|
not modelled |
50.1 |
20 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 72 | c3on5B_ |
|
not modelled |
50.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bh1974 protein;
PDBTitle: crystal structure of a xanthine dehydrogenase (bh1974) from bacillus2 halodurans at 2.80 a resolution
|
| 73 | d1u6ea2 |
|
not modelled |
49.8 |
15 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 74 | c3iupB_ |
|
not modelled |
49.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadph:quinone oxidoreductase;
PDBTitle: crystal structure of putative nadph:quinone oxidoreductase2 (yp_296108.1) from ralstonia eutropha jmp134 at 1.70 a resolution
|
| 75 | c3h75A_ |
|
not modelled |
49.5 |
11 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic sugar-binding domain protein;
PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
|
| 76 | c3nrnA_ |
|
not modelled |
48.9 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pf1083;
PDBTitle: crystal structure of pf1083 protein from pyrococcus furiosus,2 northeast structural genomics consortium target pfr223
|
| 77 | c3jskN_ |
|
not modelled |
48.8 |
8 |
PDB header:biosynthetic protein
Chain: N: PDB Molecule:cypbp37 protein;
PDBTitle: thiazole synthase from neurospora crassa
|
| 78 | c2vdcI_ |
|
not modelled |
48.1 |
14 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 79 | c1sezA_ |
|
not modelled |
48.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase, mitochondrial;
PDBTitle: crystal structure of protoporphyrinogen ix oxidase
|
| 80 | d2h00a1 |
|
not modelled |
47.8 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Methyltransferase 10 domain |
| 81 | d1lpfa1 |
|
not modelled |
47.8 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 82 | d2gf3a1 |
|
not modelled |
47.4 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 83 | c1vi2B_ |
|
not modelled |
47.3 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase 2;
PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
|
| 84 | c3kljA_ |
|
not modelled |
47.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-dependent dehydrogenase, nirb-family (n-terminal
PDBTitle: crystal structure of nadh:rubredoxin oxidoreductase from clostridium2 acetobutylicum
|
| 85 | d1o8bb1 |
|
not modelled |
47.2 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 86 | d1otha2 |
|
not modelled |
46.9 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 87 | d1neka2 |
|
not modelled |
46.6 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 88 | c3fbsB_ |
|
not modelled |
46.2 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of the oxidoreductase from agrobacterium2 tumefaciens
|
| 89 | d2cula1 |
|
not modelled |
46.2 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GidA-like |
| 90 | c3k30B_ |
|
not modelled |
46.1 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 91 | c3ma0A_ |
|
not modelled |
46.0 |
11 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:d-xylose-binding periplasmic protein;
PDBTitle: closed liganded crystal structure of xylose binding protein from2 escherichia coli
|
| 92 | c3f8rD_ |
|
not modelled |
45.9 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase (trxb-3);
PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
|
| 93 | d1k0ia1 |
|
not modelled |
44.2 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 94 | c3muxB_ |
|
not modelled |
44.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
| 95 | d1v59a1 |
|
not modelled |
44.0 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 96 | c2j37W_ |
|
not modelled |
43.8 |
13 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein
PDBTitle: model of mammalian srp bound to 80s rncs
|
| 97 | c1txtB_ |
|
not modelled |
43.8 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:3-hydroxy-3-methylglutaryl-coa synthase;
PDBTitle: staphylococcus aureus 3-hydroxy-3-methylglutaryl-coa2 synthase
|
| 98 | d1hnja2 |
|
not modelled |
42.7 |
20 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 99 | c3v76A_ |
|
not modelled |
42.7 |
21 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavoprotein;
PDBTitle: the crystal structure of a flavoprotein from sinorhizobium meliloti
|
| 100 | c1f8sA_ |
|
not modelled |
42.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-amino acid oxidase;
PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
|
| 101 | c3g05B_ |
|
not modelled |
42.3 |
26 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of n-terminal domain (2-550) of e.coli mnmg
|
| 102 | c3cgvA_ |
|
not modelled |
42.2 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:geranylgeranyl reductase related protein;
PDBTitle: crystal structure of geranylgeranyl bacteriochlorophyll reductase-like2 fixc homolog (np_393992.1) from thermoplasma acidophilum at 1.60 a3 resolution
|
| 103 | d1gesa1 |
|
not modelled |
42.1 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 104 | c2gqfA_ |
|
not modelled |
42.0 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0933;
PDBTitle: crystal structure of flavoprotein hi0933 from haemophilus influenzae2 rd
|
| 105 | c2xdoC_ |
|
not modelled |
41.8 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:tetx2 protein;
PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
|
| 106 | c1ps9A_ |
|
not modelled |
41.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 107 | d2bs2a2 |
|
not modelled |
41.5 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 108 | d1djqa3 |
|
not modelled |
41.4 |
19 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 109 | c2bryA_ |
|
not modelled |
41.4 |
24 |
PDB header:transport
Chain: A: PDB Molecule:nedd9 interacting protein with calponin homology
PDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
|
| 110 | c1mzjB_ |
|
not modelled |
41.2 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:beta-ketoacylsynthase iii;
PDBTitle: crystal structure of the priming beta-ketosynthase from the2 r1128 polyketide biosynthetic pathway
|
| 111 | d2voua1 |
|
not modelled |
41.2 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 112 | d1mzja2 |
|
not modelled |
41.0 |
19 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 113 | d1h6va1 |
|
not modelled |
40.4 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 114 | c2rghA_ |
|
not modelled |
40.1 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 115 | d1onfa1 |
|
not modelled |
39.9 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 116 | d1bg6a2 |
|
not modelled |
39.9 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 117 | c1yvvB_ |
|
not modelled |
39.9 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase, flavin-containing;
PDBTitle: x-ray structurure of p. syringae q888a4 oxidoreductase at2 resolution 2.5a. northeast structural genomics consortium3 target psr10.
|
| 118 | d1mv8a2 |
|
not modelled |
39.9 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 119 | d2g5ca2 |
|
not modelled |
39.6 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 120 | c2i0zA_ |
|
not modelled |
39.6 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-utilizing dehydrogenases;
PDBTitle: crystal structure of a fad binding protein from bacillus2 cereus, a putative nad(fad)-utilizing dehydrogenases
|

