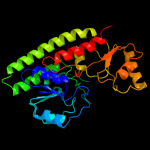
| 1 | c2ps3A_ |
|
 |
100.0 |
99 |
PDB header:metal transport
Chain: A: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: structure and metal binding properties of znua, a2 periplasmic zinc transporter from escherichia coli
|
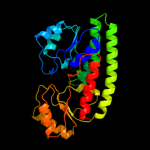
| 2 | c2ogwB_ |
|
 |
100.0 |
97 |
PDB header:transport protein
Chain: B: PDB Molecule:high-affinity zinc uptake system protein znua
PDBTitle: structure of abc type zinc transporter from e. coli
|
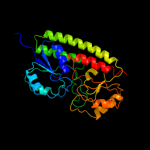
| 3 | d1xvla1 |
|
 |
100.0 |
20 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
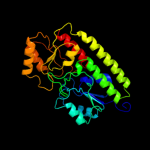
| 4 | d1psza_ |
|
 |
100.0 |
20 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
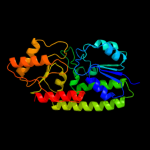
| 5 | c1toaA_ |
|
 |
100.0 |
21 |
PDB header:binding protein
Chain: A: PDB Molecule:protein (periplasmic binding protein troa);
PDBTitle: periplasmic zinc binding protein troa from treponema pallidum
|
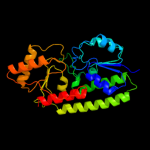
| 6 | d1toaa_ |
|
 |
100.0 |
21 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
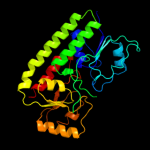
| 7 | c3cx3A_ |
|
 |
100.0 |
23 |
PDB header:metal binding protein
Chain: A: PDB Molecule:lipoprotein;
PDBTitle: crystal structure analysis of the streptococcus pneumoniae2 adcaii protein
|
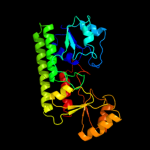
| 8 | c2o1eB_ |
|
 |
100.0 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:ycdh;
PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
|
| 9 | c3hjtB_ |
|
 |
100.0 |
25 |
PDB header:cell adhesion, transport protein
Chain: B: PDB Molecule:lmb;
PDBTitle: structure of laminin binding protein (lmb) of streptococcus2 agalactiae a bifunctional protein with adhesin and metal3 transporting activity
|
| 10 | c3mfqB_ |
|
 |
100.0 |
20 |
PDB header:metal binding protein
Chain: B: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: a glance into the metal binding specificity of troa: where elaborate2 behaviors occur in the active center
|
| 11 | c2ov3A_ |
|
 |
100.0 |
26 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein component of an abc
PDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc2 bound
|
| 12 | d1pq4a_ |
|
 |
100.0 |
27 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
| 13 | c1xvlC_ |
|
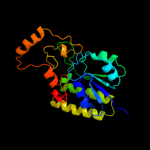 |
100.0 |
21 |
PDB header:metal transport
Chain: C: PDB Molecule:mn transporter;
PDBTitle: the three-dimensional structure of mntc from synechocystis2 6803
|
| 14 | d1qh8a_ |
|
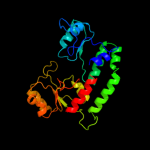 |
97.2 |
8 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 15 | c2xdqB_ |
|
 |
96.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 16 | c3pdiG_ |
|
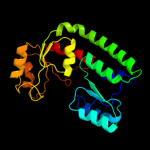 |
96.0 |
14 |
PDB header:protein binding
Chain: G: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nife;
PDBTitle: precursor bound nifen
|
| 17 | d1mioa_ |
|
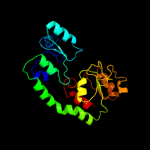 |
95.7 |
20 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 18 | c3pdiB_ |
|
 |
95.6 |
15 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
| 19 | d1m1na_ |
|
 |
94.9 |
11 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 20 | d1miob_ |
|
 |
92.9 |
18 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 21 | c3aerB_ |
|
not modelled |
92.7 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
|
| 22 | d2ez9a1 |
|
not modelled |
91.9 |
20 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 23 | c2gejA_ |
|
not modelled |
89.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol mannosyltransferase (pima);
PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
|
| 24 | c3c4vB_ |
|
not modelled |
88.3 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:predicted glycosyltransferases;
PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
|
| 25 | d1qh8b_ |
|
not modelled |
88.3 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 26 | d1m1nb_ |
|
not modelled |
87.7 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 27 | c2r60A_ |
|
not modelled |
79.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 28 | d2iw1a1 |
|
not modelled |
78.7 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 29 | c3s29C_ |
|
not modelled |
74.8 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:sucrose synthase 1;
PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
|
| 30 | d1a9xa2 |
|
not modelled |
70.6 |
16 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 31 | c3lhkA_ |
|
not modelled |
70.3 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative dna binding protein mj0014;
PDBTitle: crystal structure of putative dna binding protein from2 methanocaldococcus jannaschii.
|
| 32 | d2ji7a1 |
|
not modelled |
70.1 |
9 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 33 | d1uqta_ |
|
not modelled |
65.1 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Trehalose-6-phosphate synthase, OtsA |
| 34 | c3ff4A_ |
|
not modelled |
64.7 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
| 35 | c2jjmH_ |
|
not modelled |
63.5 |
10 |
PDB header:transferase
Chain: H: PDB Molecule:glycosyl transferase, group 1 family protein;
PDBTitle: crystal structure of a family gt4 glycosyltransferase from2 bacillus anthracis orf ba1558.
|
| 36 | d1iuka_ |
|
not modelled |
63.3 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 37 | c3aerC_ |
|
not modelled |
63.0 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
|
| 38 | c1uquB_ |
|
not modelled |
61.0 |
13 |
PDB header:synthase
Chain: B: PDB Molecule:alpha, alpha-trehalose-phosphate synthase;
PDBTitle: trehalose-6-phosphate from e. coli bound with udp-glucose.
|
| 39 | d2bisa1 |
|
not modelled |
60.6 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 40 | c2xdqA_ |
|
not modelled |
60.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 41 | c3gk0H_ |
|
not modelled |
59.9 |
19 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
|
| 42 | c2qjhH_ |
|
not modelled |
59.6 |
8 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 43 | d1i60a_ |
|
not modelled |
57.4 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 44 | c3lmzA_ |
|
not modelled |
57.1 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
| 45 | c3o6cA_ |
|
not modelled |
56.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
|
| 46 | d1xg7a_ |
|
not modelled |
54.6 |
11 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:AgoG-like |
| 47 | d1wo8a1 |
|
not modelled |
54.0 |
16 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 48 | c2qzsA_ |
|
not modelled |
54.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:glycogen synthase;
PDBTitle: crystal structure of wild-type e.coli gs in complex with adp2 and glucose(wtgsb)
|
| 49 | c3dx5A_ |
|
not modelled |
53.9 |
6 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 50 | c3shoA_ |
|
not modelled |
53.2 |
10 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, rpir family;
PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
|
| 51 | c2xhzC_ |
|
not modelled |
53.1 |
10 |
PDB header:isomerase
Chain: C: PDB Molecule:arabinose 5-phosphate isomerase;
PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
|
| 52 | d1x94a_ |
|
not modelled |
52.1 |
8 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 53 | c3uhjE_ |
|
not modelled |
51.5 |
10 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:probable glycerol dehydrogenase;
PDBTitle: crystal structure of a probable glycerol dehydrogenase from2 sinorhizobium meliloti 1021
|
| 54 | d1x9ma1 |
|
not modelled |
50.3 |
11 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:DnaQ-like 3'-5' exonuclease |
| 55 | c2a3nA_ |
|
not modelled |
49.9 |
8 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative glucosamine-fructose-6-phosphate aminotransferase;
PDBTitle: crystal structure of a putative glucosamine-fructose-6-phosphate2 aminotransferase (stm4540.s) from salmonella typhimurium lt2 at 1.353 a resolution
|
| 56 | d2djia1 |
|
not modelled |
49.8 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 57 | c3a24A_ |
|
not modelled |
49.2 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of bt1871 retaining glycosidase
|
| 58 | c2xecD_ |
|
not modelled |
48.9 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 59 | d1qcza_ |
|
not modelled |
46.0 |
11 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 60 | c2duwA_ |
|
not modelled |
45.2 |
7 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:putative coa-binding protein;
PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
|
| 61 | d1m6ja_ |
|
not modelled |
44.9 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 62 | d2f9fa1 |
|
not modelled |
44.3 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 63 | d2hk6a1 |
|
not modelled |
43.8 |
17 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 64 | d2chua1 |
|
not modelled |
43.6 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TM0189-like |
| 65 | c3orsD_ |
|
not modelled |
43.4 |
13 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
| 66 | d1ybha1 |
|
not modelled |
42.9 |
11 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 67 | c3trhI_ |
|
not modelled |
42.6 |
11 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
| 68 | c3o3nA_ |
|
not modelled |
42.3 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:alpha-subunit 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 69 | d1qtwa_ |
|
not modelled |
42.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 70 | c3s99A_ |
|
not modelled |
40.6 |
11 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
| 71 | c3iwpK_ |
|
not modelled |
40.4 |
10 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
| 72 | c2yvqA_ |
|
not modelled |
39.6 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 73 | c3okaA_ |
|
not modelled |
39.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:gdp-mannose-dependent alpha-(1-6)-phosphatidylinositol
PDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
|
| 74 | c3rhzB_ |
|
not modelled |
39.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:nucleotide sugar synthetase-like protein;
PDBTitle: structure and functional analysis of a new subfamily of2 glycosyltransferases required for glycosylation of serine-rich3 streptococcal adhesions
|
| 75 | c2q8pA_ |
|
not modelled |
38.8 |
12 |
PDB header:metal transport
Chain: A: PDB Molecule:iron-regulated surface determinant e;
PDBTitle: crystal structure of selenomethionine labelled s. aureus isde2 complexed with heme
|
| 76 | d3bc8a1 |
|
not modelled |
38.2 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
| 77 | d1m5wa_ |
|
not modelled |
37.4 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:Pyridoxine 5'-phosphate synthase
Family:Pyridoxine 5'-phosphate synthase |
| 78 | c3kwsB_ |
|
not modelled |
36.3 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 79 | c3trjC_ |
|
not modelled |
35.6 |
8 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
| 80 | d2q02a1 |
|
not modelled |
35.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 81 | c2k6xA_ |
|
not modelled |
35.3 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
|
| 82 | d1u11a_ |
|
not modelled |
35.1 |
11 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 83 | c2h9aA_ |
|
not modelled |
34.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 84 | d1jq5a_ |
|
not modelled |
34.3 |
11 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 85 | d1pvva1 |
|
not modelled |
33.7 |
18 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 86 | c2x0dA_ |
|
not modelled |
33.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:wsaf;
PDBTitle: apo structure of wsaf
|
| 87 | c3qxbB_ |
|
not modelled |
32.9 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
| 88 | d1xmpa_ |
|
not modelled |
32.6 |
11 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 89 | c3hl2D_ |
|
not modelled |
32.4 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:o-phosphoseryl-trna(sec) selenium transferase;
PDBTitle: the crystal structure of the human sepsecs-trnasec complex
|
| 90 | d1t9ba1 |
|
not modelled |
32.3 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 91 | c2fw9A_ |
|
not modelled |
32.1 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
| 92 | d1duvg1 |
|
not modelled |
31.2 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 93 | c1u8cB_ |
|
not modelled |
30.6 |
8 |
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
|
| 94 | c1mgpA_ |
|
not modelled |
30.4 |
10 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:hypothetical protein tm841;
PDBTitle: hypothetical protein tm841 from thermotoga maritima reveals2 fatty acid binding function
|
| 95 | d1mgpa_ |
|
not modelled |
30.4 |
10 |
Fold:DAK1/DegV-like
Superfamily:DAK1/DegV-like
Family:DegV-like |
| 96 | d1aw1a_ |
|
not modelled |
29.7 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 97 | c2hqbA_ |
|
not modelled |
29.7 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional activator of comk gene;
PDBTitle: crystal structure of a transcriptional activator of comk2 gene from bacillus halodurans
|
| 98 | d1p8aa_ |
|
not modelled |
29.1 |
20 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 99 | c3o3nB_ |
|
not modelled |
28.9 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:beta-subunit 2-hydroxyacyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 100 | c3louB_ |
|
not modelled |
28.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 101 | c2ou4C_ |
|
not modelled |
28.7 |
6 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 102 | d1o4va_ |
|
not modelled |
28.6 |
16 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 103 | d2csua2 |
|
not modelled |
28.1 |
16 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 104 | c1ezaA_ |
|
not modelled |
28.1 |
12 |
PDB header:phosphotransferase
Chain: A: PDB Molecule:enzyme i;
PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
|
| 105 | d1y81a1 |
|
not modelled |
28.0 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 106 | d1ub3a_ |
|
not modelled |
27.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 107 | c3nbmA_ |
|
not modelled |
27.7 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 108 | d2bfwa1 |
|
not modelled |
27.1 |
18 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 109 | c1ortD_ |
|
not modelled |
27.1 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine transcarbamoylase;
PDBTitle: ornithine transcarbamoylase from pseudomonas aeruginosa
|
| 110 | c3g9qA_ |
|
not modelled |
27.0 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:ferrichrome-binding protein;
PDBTitle: crystal structure of the fhud fold-family bsu3320, a periplasmic2 binding protein component of a fep/fec-like ferrichrome abc3 transporter from bacillus subtilis. northeast structural genomics4 consortium target sr577a
|
| 111 | c3lp6D_ |
|
not modelled |
26.7 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
| 112 | d1u7pa_ |
|
not modelled |
26.4 |
7 |
Fold:HAD-like
Superfamily:HAD-like
Family:Magnesium-dependent phosphatase-1, Mdp1 |
| 113 | d1txja_ |
|
not modelled |
26.1 |
11 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:Translationally controlled tumor protein TCTP (histamine-releasing factor) |
| 114 | c3h3eA_ |
|
not modelled |
26.1 |
9 |
PDB header:structural genomics, metal binding prote
Chain: A: PDB Molecule:uncharacterized protein tm1679;
PDBTitle: crystal structure of tm1679, a metal-dependent hydrolase of2 the beta-lactamase superfamily
|
| 115 | c1vlvA_ |
|
not modelled |
25.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
|
| 116 | c2cazB_ |
|
not modelled |
25.4 |
9 |
PDB header:protein transport
Chain: B: PDB Molecule:vacuolar protein sorting-associated protein
PDBTitle: escrt-i core
|
| 117 | d2cazb1 |
|
not modelled |
25.4 |
9 |
Fold:Long alpha-hairpin
Superfamily:Endosomal sorting complex assembly domain
Family:VPS28 N-terminal domain |
| 118 | d2bb3a1 |
|
not modelled |
25.2 |
7 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 119 | d1nu0a_ |
|
not modelled |
25.0 |
10 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 120 | c2qniA_ |
|
not modelled |
24.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu0299;
PDBTitle: crystal structure of uncharacterized protein atu0299
|

