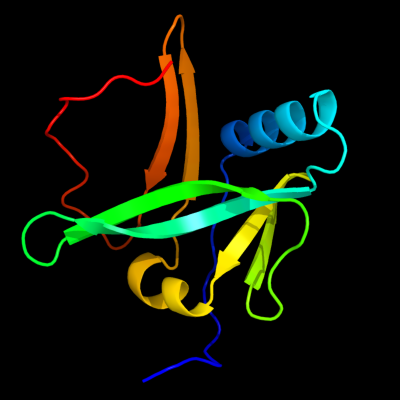
| 1 | c2d2aA_
|
|
 |
100.0 |
100 |
PDB header:metal transport
Chain: A: PDB Molecule:sufa protein;
PDBTitle: crystal structure of escherichia coli sufa involved in2 biosynthesis of iron-sulfur clusters
|
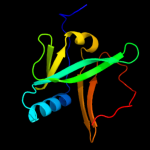
| 2 | c2apnA_
|
|
 |
100.0 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein hi1723;
PDBTitle: hi1723 solution structure
|
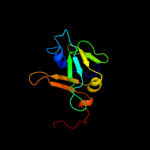
| 3 | c1x0gA_
|
|
 |
100.0 |
35 |
PDB header:metal binding protein
Chain: A: PDB Molecule:isca;
PDBTitle: crystal structure of isca with the [2fe-2s] cluster
|
| 4 | d1s98a_
|
|
 |
100.0 |
44 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 5 | c2k4zA_
|
|
 |
100.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsrr;
PDBTitle: solution nmr structure of allochromatium vinosum dsrr:2 northeast structural genomics consortium target op5
|
| 6 | d1nwba_
|
|
 |
99.9 |
26 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 7 | d2p2ea1
|
|
 |
98.7 |
14 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 8 | c2qgoA_
|
|
 |
98.4 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative fe-s biosynthesis protein;
PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
|
| 9 | c3pm7A_
|
|
 |
33.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of ef_3132 protein from enterococcus faecalis at the2 resolution 2a, northeast structural genomics consortium target efr184
|
| 10 | d2frha1
|
|
 |
23.0 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 11 | d1sp2a_
|
|
 |
21.1 |
44 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 12 | d1zfda_
|
|
 |
18.1 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 13 | d1a1ia1
|
|
 |
16.5 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 14 | d2glia3
|
|
 |
16.4 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 15 | d1aaya1
|
|
 |
15.6 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 16 | d1p4xa1
|
|
 |
15.6 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 17 | d1ubdc3
|
|
 |
15.2 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 18 | d1p4xa2
|
|
 |
15.2 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 19 | c3obhA_
|
|
 |
14.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: x-ray crystal structure of protein sp_0782 (7-79) from streptococcus2 pneumoniae. northeast structural genomics consortium target spr104
|
| 20 | d1a1ha1
|
|
 |
14.7 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 21 | d1f2ig1 |
|
not modelled |
14.6 |
33 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 22 | d1hsja1 |
|
not modelled |
14.2 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 23 | d1a1ga1 |
|
not modelled |
14.2 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 24 | c2kvfA_ |
|
not modelled |
14.1 |
38 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger and btb domain-containing protein 32;
PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
|
| 25 | c1p4xA_ |
|
not modelled |
13.8 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:staphylococcal accessory regulator a homologue;
PDBTitle: crystal structure of sars protein from staphylococcus aureus
|
| 26 | d1tf3a2 |
|
not modelled |
13.8 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 27 | d1u86a1 |
|
not modelled |
11.7 |
44 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 28 | d1vsra_ |
|
not modelled |
10.7 |
17 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Very short patch repair (VSR) endonuclease |
| 29 | d1l7da2 |
|
not modelled |
10.0 |
20 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:L-alanine dehydrogenase-like |
| 30 | d1ncsa_ |
|
not modelled |
10.0 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 31 | d2dlka2 |
|
not modelled |
9.7 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 32 | c1bcrA_ |
|
not modelled |
9.7 |
15 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
|
| 33 | d2yt9a1 |
|
not modelled |
9.6 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 34 | d2h85a2 |
|
not modelled |
9.5 |
29 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 35 | c3me5A_ |
|
not modelled |
9.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:cytosine-specific methyltransferase;
PDBTitle: crystal structure of putative dna cytosine methylase from shigella2 flexneri 2a str. 301
|
| 36 | c2kdxA_ |
|
not modelled |
9.3 |
26 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
| 37 | c2ab3A_ |
|
not modelled |
9.3 |
67 |
PDB header:rna binding protein
Chain: A: PDB Molecule:znf29;
PDBTitle: solution structures and characterization of hiv rre iib rna2 targeting zinc finger proteins
|
| 38 | d1x6ea2 |
|
not modelled |
9.0 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 39 | c2yujA_ |
|
not modelled |
9.0 |
25 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
|
| 40 | d1rp0a1 |
|
not modelled |
8.8 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Thi4-like |
| 41 | c1zc1A_ |
|
not modelled |
8.5 |
23 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
| 42 | c3a44D_ |
|
not modelled |
8.3 |
17 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
| 43 | c1va1A_ |
|
not modelled |
8.2 |
57 |
PDB header:transcription
Chain: A: PDB Molecule:transcription factor sp1;
PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 1)
|
| 44 | d1jj7a_ |
|
not modelled |
8.0 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 45 | c3c3jA_ |
|
not modelled |
7.9 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
|
| 46 | c1znmA_ |
|
not modelled |
7.8 |
38 |
PDB header:zinc finger
Chain: A: PDB Molecule:yy1;
PDBTitle: a zinc finger with an artificial beta-turn, original2 sequence taken from the third zinc finger domain of the3 human transcriptional repressor protein yy1 (ying and yang4 1, a delta transcription factor), nmr, 34 structures
|
| 47 | d1wgea1 |
|
not modelled |
7.7 |
40 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 48 | c3odpA_ |
|
not modelled |
7.1 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
|
| 49 | c2qrlA_ |
|
not modelled |
7.1 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase, nad+, l-lysine-
PDBTitle: crystal structure of oxalylglycine-bound saccharopine2 dehydrogenase (l-lys forming) from saccharomyces cerevisiae
|
| 50 | d1wmia1 |
|
not modelled |
7.1 |
7 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
| 51 | c2nq2C_ |
|
not modelled |
7.0 |
16 |
PDB header:metal transport
Chain: C: PDB Molecule:hypothetical abc transporter atp-binding protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
|
| 52 | c3g5oC_ |
|
not modelled |
6.9 |
20 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
| 53 | c1gxsC_ |
|
not modelled |
6.7 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 54 | d2cota1 |
|
not modelled |
6.6 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 55 | c3nbbC_ |
|
not modelled |
6.5 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:peroxisomal primary amine oxidase;
PDBTitle: crystal structure of mutant y305f expressed in e. coli in the copper2 amine oxidase from hansenula polymorpha
|
| 56 | c2entA_ |
|
not modelled |
6.0 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:krueppel-like factor 15;
PDBTitle: solution structure of the second c2h2-type zinc finger2 domain from human krueppel-like factor 15
|
| 57 | c1ekmC_ |
|
not modelled |
5.8 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure at 2.5 a resolution of zinc-substituted2 copper amine oxidase of hansenula polymorpha expressed in3 escherichia coli
|
| 58 | d2bs2b2 |
|
not modelled |
5.8 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 59 | d2fbia1 |
|
not modelled |
5.7 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 60 | d1ywsa1 |
|
not modelled |
5.7 |
40 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 61 | d1qabe_ |
|
not modelled |
5.6 |
24 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 62 | d1kt7a_ |
|
not modelled |
5.5 |
16 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 63 | d1jgsa_ |
|
not modelled |
5.5 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 64 | d2cota2 |
|
not modelled |
5.4 |
67 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 65 | c2ayuA_ |
|
not modelled |
5.4 |
10 |
PDB header:chaperone
Chain: A: PDB Molecule:nucleosome assembly protein;
PDBTitle: the structure of nucleosome assembly protein suggests a mechanism for2 histone binding and shuttling
|
| 66 | d2ayua1 |
|
not modelled |
5.4 |
10 |
Fold:NAP-like
Superfamily:NAP-like
Family:NAP-like |
| 67 | c3g3zA_ |
|
not modelled |
5.3 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
|
| 68 | d1a1ia3 |
|
not modelled |
5.3 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 69 | c3h5jA_ |
|
not modelled |
5.2 |
60 |
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
|
| 70 | c3o2eA_ |
|
not modelled |
5.1 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:bola-like protein;
PDBTitle: crystal structure of a bol-like protein from babesia bovis
|
| 71 | d1u85a1 |
|
not modelled |
5.1 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |