1 c3cfuA_
63.9
14
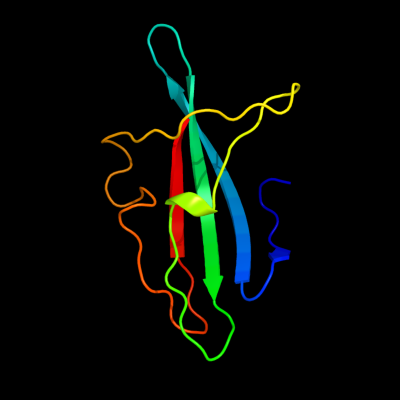
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yjha;PDBTitle: crystal structure of the yjha protein from bacillus2 subtilis. northeast structural genomics consortium target3 sr562
2 c3mk7K_
51.6
12
PDB header: oxidoreductaseChain: K: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit n;PDBTitle: the structure of cbb3 cytochrome oxidase
3 d2affa1
47.6
28
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain4 c1r21A_
46.8
30
PDB header: cell cycleChain: A: PDB Molecule: antigen ki-67;PDBTitle: solution structure of human ki67 fha domain
5 c3rkoN_
46.7
13
PDB header: oxidoreductaseChain: N: PDB Molecule: nadh-quinone oxidoreductase subunit n;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
6 c3lkxB_
29.6
25
PDB header: chaperoneChain: B: PDB Molecule: nascent polypeptide-associated complex subunit alpha;PDBTitle: human nac dimerization domain
7 c2jp3A_
29.3
29
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
8 c1agqB_
27.3
42
PDB header: growth factorChain: B: PDB Molecule: glial cell-derived neurotrophic factor;PDBTitle: glial cell-derived neurotrophic factor from rat
9 d1udxa3
26.2
29
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain10 d2ff4a3
20.9
29
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain11 d3b55a1
18.1
13
Fold: EreA/ChaN-likeSuperfamily: EreA/ChaN-likeFamily: EreA-like12 c2radB_
17.4
13
PDB header: biosynthetic proteinChain: B: PDB Molecule: succinoglycan biosynthesis protein;PDBTitle: crystal structure of the succinoglycan biosynthesis2 protein. northeast structural genomics consortium target3 bcr135
13 c1lcyA_
16.7
14
PDB header: hydrolaseChain: A: PDB Molecule: htra2 serine protease;PDBTitle: crystal structure of the mitochondrial serine protease htra2
14 c3gqsB_
14.8
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: adenylate cyclase-like protein;PDBTitle: crystal structure of the fha domain of ct664 protein from chlamydia2 trachomatis
15 c1bhbA_
14.0
31
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
16 d1dlpa1
13.9
19
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins17 c3rkoM_
12.5
15
PDB header: oxidoreductaseChain: M: PDB Molecule: nadh-quinone oxidoreductase subunit m;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
18 c2zpmA_
11.9
19
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
19 d1l9bm_
11.5
23
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits20 c3seeA_
11.4
21
PDB header: sugar binding proteinChain: A: PDB Molecule: hypothetical sugar binding protein;PDBTitle: crystal structure of a hypothetical sugar binding protein (bt_4411)2 from bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
21 c2kklA_
not modelled
10.2
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mb1858;PDBTitle: solution nmr structure of fha domain of mb1858 from2 mycobacterium bovis. northeast structural genomics3 consortium target mbr243c (24-155).
22 c3poaA_
not modelled
10.0
14
PDB header: peptide binding proteinChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: structural and functional analysis of phosphothreonine-dependent fha2 domain interactions
23 c2l9uA_
not modelled
9.9
31
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
24 d2o3ga1
not modelled
9.9
27
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like25 d2j8cm1
not modelled
9.8
23
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits26 d2f69a1
not modelled
9.6
20
Fold: open-sided beta-meanderSuperfamily: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domainFamily: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain27 c2r76A_
not modelled
9.6
15
PDB header: lipoproteinChain: A: PDB Molecule: rare lipoprotein b;PDBTitle: crystal structure of the rare lipoprotein b (so_1173) from shewanella2 oneidensis, northeast structural genomics consortium target sor91a
28 d2i4sa1
not modelled
9.4
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like29 c3pv4A_
not modelled
9.3
33
PDB header: hydrolaseChain: A: PDB Molecule: degq;PDBTitle: structure of legionella fallonii degq (delta-pdz2 variant)
30 d3dtua1
not modelled
9.1
11
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like31 c2jo1A_
not modelled
8.9
19
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
32 d2bgca2
not modelled
8.8
80
Fold: Double-stranded beta-helixSuperfamily: cAMP-binding domain-likeFamily: Listeriolysin regulatory protein PrfA, N-terminal domain33 d1hdmb2
not modelled
8.7
83
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain34 c4a0kB_
not modelled
8.6
75
PDB header: ligase/dna-binding protein/dnaChain: B: PDB Molecule: e3 ubiquitin-protein ligase rbx1;PDBTitle: structure of ddb1-ddb2-cul4a-rbx1 bound to a 12 bp abasic2 site containing dna-duplex
35 c3uotB_
not modelled
8.5
7
PDB header: cell cycleChain: B: PDB Molecule: mediator of dna damage checkpoint protein 1;PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
36 c2lezA_
not modelled
8.5
20
PDB header: signaling proteinChain: A: PDB Molecule: secreted effector protein pipb2;PDBTitle: solution nmr structure of n-terminal domain of salmonella effector2 protein pipb2. northeast structural genomics consortium (nesg) target3 stt318a
37 d2i6va1
not modelled
8.4
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like38 d1x9la_
not modelled
8.3
13
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: DR1885-like metal-binding proteinFamily: DR1885-like metal-binding protein39 d1rl2a2
not modelled
8.1
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like40 d2p13a1
not modelled
8.0
16
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like41 c2q04C_
not modelled
7.9
43
PDB header: transferaseChain: C: PDB Molecule: acetoin utilization protein;PDBTitle: crystal structure of acetoin utilization protein (zp_00540088.1) from2 exiguobacterium sibiricum 255-15 at 2.33 a resolution
42 c2kjpA_
not modelled
7.8
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ylbl;PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
43 c1yj5C_
not modelled
7.5
7
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
44 c3ia8A_
not modelled
7.5
29
PDB header: metal binding proteinChain: A: PDB Molecule: thap domain-containing protein 4;PDBTitle: the structure of the c-terminal heme nitrobindin domain of thap2 domain-containing protein 4 from homo sapiens
45 c2p3wB_
not modelled
7.4
17
PDB header: protein bindingChain: B: PDB Molecule: probable serine protease htra3;PDBTitle: crystal structure of the htra3 pdz domain bound to a phage-derived2 ligand (fgrwv)
46 c3htxA_
not modelled
7.3
19
PDB header: transferase/rnaChain: A: PDB Molecule: hen1;PDBTitle: crystal structure of small rna methyltransferase hen1
47 c1hgvA_
not modelled
7.3
21
PDB header: virusChain: A: PDB Molecule: ph75 inovirus major coat protein;PDBTitle: filamentous bacteriophage ph75
48 d2qgma1
not modelled
7.3
13
Fold: EreA/ChaN-likeSuperfamily: EreA/ChaN-likeFamily: EreA-like49 c2qgmA_
not modelled
7.3
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: succinoglycan biosynthesis protein;PDBTitle: crystal structure of succinoglycan biosynthesis protein at2 the resolution 1.7 a. northeast structural genomics3 consortium target bcr136.
50 d1eysm_
not modelled
7.3
18
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits51 c3rkoL_
not modelled
7.3
13
PDB header: oxidoreductaseChain: L: PDB Molecule: nadh-quinone oxidoreductase subunit l;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
52 c3mpbA_
not modelled
7.2
14
PDB header: isomeraseChain: A: PDB Molecule: sugar isomerase;PDBTitle: z5688 from e. coli o157:h7 bound to fructose
53 d1gh6b2
not modelled
7.2
27
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Retinoblastoma tumor suppressor domains54 c3i18A_
not modelled
7.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2051 protein;PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
55 c3stjC_
not modelled
7.1
24
PDB header: hydrolaseChain: C: PDB Molecule: protease degq;PDBTitle: crystal structure of the protease + pdz1 domain of degq from2 escherichia coli
56 c3eh4A_
not modelled
7.0
13
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase subunit 1;PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
57 c1sddB_
not modelled
6.9
17
PDB header: blood clottingChain: B: PDB Molecule: coagulation factor v;PDBTitle: crystal structure of bovine factor vai
58 c3lw52_
not modelled
6.9
21
PDB header: photosynthesisChain: 2: PDB Molecule: type ii chlorophyll a/b binding protein from photosystem i;PDB Fragment: residues 81-246;
PDBTitle: improved model of plant photosystem i
59 d1q90r_
not modelled
6.8
31
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor60 c1q90R_
not modelled
6.8
31
PDB header: photosynthesisChain: R: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
61 c1dlpA_
not modelled
6.8
19
PDB header: sugar binding proteinChain: A: PDB Molecule: lectin scafet precursor;PDBTitle: structural characterization of the native fetuin-binding2 protein scilla campanulata agglutinin (scafet): a novel3 two-domain lectin
62 d1e57a_
not modelled
6.7
29
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Tymoviridae-like VP63 d1r45a_
not modelled
6.6
11
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins64 c1zrtD_
not modelled
6.6
10
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
65 d1uapa_
not modelled
6.6
15
Fold: OB-foldSuperfamily: TIMP-likeFamily: Netrin-like domain (NTR/C345C module)66 c2yvxD_
not modelled
6.5
18
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
67 d1qbaa2
not modelled
6.4
9
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Bacterial chitobiase, n-terminal domain68 d1vqoa2
not modelled
6.3
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like69 c2rf9D_
not modelled
6.3
43
PDB header: transferaseChain: D: PDB Molecule: erbb receptor feedback inhibitor 1;PDBTitle: crystal structure of the complex between the egfr kinase2 domain and a mig6 peptide
70 d1i32a2
not modelled
6.3
18
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like71 d1ggaa2
not modelled
6.3
24
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like72 d2r7ga2
not modelled
6.3
27
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Retinoblastoma tumor suppressor domains73 d2z9ia1
not modelled
6.2
11
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases74 d2o1ra1
not modelled
6.2
21
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like75 d2plia1
not modelled
6.2
23
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like76 c2kl1A_
not modelled
6.2
18
PDB header: protein bindingChain: A: PDB Molecule: ylbl protein;PDBTitle: solution structure of gtr34c from geobacillus thermodenitrificans.2 northeast structural genomics consortium target gtr34c
77 c2bbjB_
not modelled
6.1
12
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
78 d2pkqo2
not modelled
6.1
18
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like79 d2brfa1
not modelled
6.1
8
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain80 d1t3la2
not modelled
6.1
36
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases81 c3llbA_
not modelled
6.1
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
82 d1q90n_
not modelled
6.1
35
Fold: Single transmembrane helixSuperfamily: PetN subunit of the cytochrome b6f complexFamily: PetN subunit of the cytochrome b6f complex83 d2c8aa1
not modelled
6.1
11
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins84 d1vjja2
not modelled
6.0
8
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Transglutaminase, two C-terminal domainsFamily: Transglutaminase, two C-terminal domains85 d3deda1
not modelled
6.0
16
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like86 c3dedB_
not modelled
6.0
16
PDB header: membrane proteinChain: B: PDB Molecule: probable hemolysin;PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
87 c1qcrD_
not modelled
5.9
19
PDB header: PDB COMPND: 88 d1uhta_
not modelled
5.9
18
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain89 d1e57b_
not modelled
5.9
29
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Tymoviridae-like VP90 c3cwbQ_
not modelled
5.9
16
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
91 c2l5bA_
not modelled
5.8
33
PDB header: apoptosisChain: A: PDB Molecule: activator of apoptosis harakiri;PDBTitle: solution structure of the transmembrane domain of bcl-2 member2 harakiri in micelles
92 d1k3ta2
not modelled
5.8
24
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like93 d2vera1
not modelled
5.8
40
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Dr-family adhesin94 d1vf5c3
not modelled
5.8
24
Fold: Single transmembrane helixSuperfamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchorFamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchor95 d1rhzb_
not modelled
5.7
24
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit96 c2bovB_
not modelled
5.7
11
PDB header: transferaseChain: B: PDB Molecule: mono-adp-ribosyltransferase c3;PDBTitle: molecular recognition of an adp-ribosylating clostridium2 botulinum c3 exoenzyme by rala gtpase
97 c3p56F_
not modelled
5.6
17
PDB header: hydrolase/replicationChain: F: PDB Molecule: ribonuclease h2 subunit c;PDBTitle: the structure of the human rnase h2 complex defines key interaction2 interfaces relevant to enzyme function and human disease
98 d1ng0c_
not modelled
5.6
38
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Tombusviridae-like VP99 d1fftc_
not modelled
5.5
7
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like