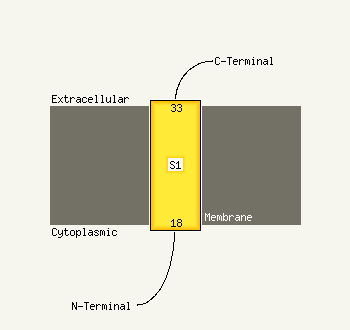
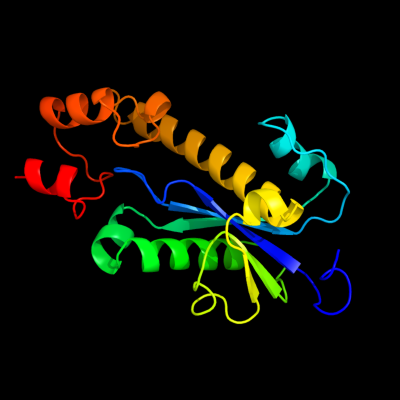
| 1 | d2etja1
|
|
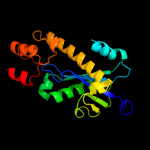 |
100.0 |
45 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 2 | c2etjA_
|
|
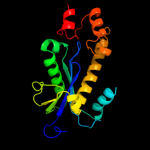 |
100.0 |
45 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: crystal structure of ribonuclease hii (ec 3.1.26.4) (rnase hii)2 (tm0915) from thermotoga maritima at 1.74 a resolution
|
| 3 | c3kioA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease h2 subunit a;
PDBTitle: mouse rnase h2 complex
|
| 4 | d1io2a_
|
|
 |
100.0 |
30 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 5 | d1uaxa_
|
|
 |
100.0 |
29 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 6 | c1i3aA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: rnase hii from archaeoglobus fulgidus with cobalt hexammine2 chloride
|
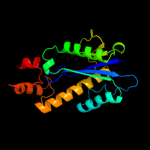
| 7 | d1i39a_
|
|
 |
100.0 |
30 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
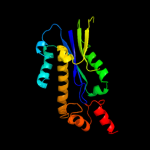
| 8 | d1ekea_
|
|
 |
100.0 |
26 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
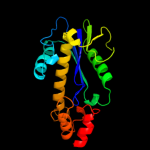
| 9 | c2d0bA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hiii;
PDBTitle: crystal structure of bst-rnase hiii in complex with mg2+
|
| 10 | c3gocB_
|
|
 |
33.1 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease v (sav1684) from streptomyces2 avermitilis. northeast structural genomics consortium target svr196
|
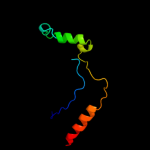
| 11 | d1c8ba_
|
|
 |
32.9 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Germination protease |
| 12 | c3c6aA_
|
|
 |
32.5 |
33 |
PDB header:viral protein
Chain: A: PDB Molecule:terminase large subunit;
PDBTitle: crystal structure of the rb49 gp17 nuclease domain
|
| 13 | c3ga2A_
|
|
 |
32.3 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease_v (bsu36170) from2 bacillus subtilis, northeast structural genomics3 consortium target sr624
|
| 14 | c2w36B_
|
|
 |
29.9 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:endonuclease v;
PDBTitle: structures of endonuclease v with dna reveal initiation of2 deaminated adenine repair
|
| 15 | c3lbwC_
|
|
 |
22.5 |
60 |
PDB header:transport protein
Chain: C: PDB Molecule:m2 protein;
PDBTitle: high resolution crystal structure of transmembrane domain of m2
|
| 16 | c3lbwA_
|
|
 |
22.5 |
60 |
PDB header:transport protein
Chain: A: PDB Molecule:m2 protein;
PDBTitle: high resolution crystal structure of transmembrane domain of m2
|
| 17 | c3lbwB_
|
|
 |
22.5 |
60 |
PDB header:transport protein
Chain: B: PDB Molecule:m2 protein;
PDBTitle: high resolution crystal structure of transmembrane domain of m2
|
| 18 | c3lbwD_
|
|
 |
22.5 |
60 |
PDB header:transport protein
Chain: D: PDB Molecule:m2 protein;
PDBTitle: high resolution crystal structure of transmembrane domain of m2
|
| 19 | c3c9jA_
|
|
 |
20.6 |
54 |
PDB header:membrane protein
Chain: A: PDB Molecule:proton channel protein m2, transmembrane segment;
PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
|
| 20 | c3c9jB_
|
|
 |
20.6 |
54 |
PDB header:membrane protein
Chain: B: PDB Molecule:proton channel protein m2, transmembrane segment;
PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
|
| 21 | d1tlha_ |
|
not modelled |
18.6 |
30 |
Fold:Anti-sigma factor AsiA
Superfamily:Anti-sigma factor AsiA
Family:Anti-sigma factor AsiA |
| 22 | c3c9jC_ |
|
not modelled |
17.7 |
54 |
PDB header:membrane protein
Chain: C: PDB Molecule:proton channel protein m2, transmembrane segment;
PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
|
| 23 | c3c9jD_ |
|
not modelled |
17.7 |
54 |
PDB header:membrane protein
Chain: D: PDB Molecule:proton channel protein m2, transmembrane segment;
PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
|
| 24 | c2zpaB_ |
|
not modelled |
15.3 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:uncharacterized protein ypfi;
PDBTitle: crystal structure of trna(met) cytidine acetyltransferase
|
| 25 | c2kqtD_ |
|
not modelled |
12.4 |
50 |
PDB header:transport protein
Chain: D: PDB Molecule:m2 protein;
PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
|
| 26 | c2kqtB_ |
|
not modelled |
12.4 |
50 |
PDB header:transport protein
Chain: B: PDB Molecule:m2 protein;
PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
|
| 27 | c2kqtA_ |
|
not modelled |
12.4 |
50 |
PDB header:transport protein
Chain: A: PDB Molecule:m2 protein;
PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
|
| 28 | c2kqtC_ |
|
not modelled |
12.4 |
50 |
PDB header:transport protein
Chain: C: PDB Molecule:m2 protein;
PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
|
| 29 | d1wfza_ |
|
not modelled |
12.4 |
16 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 30 | c1mp6A_ |
|
not modelled |
11.8 |
50 |
PDB header:membrane protein
Chain: A: PDB Molecule:matrix protein m2;
PDBTitle: structure of the transmembrane region of the m2 protein h+2 channel by solid state nmr spectroscopy
|
| 31 | c1nyjC_ |
|
not modelled |
11.8 |
50 |
PDB header:viral protein
Chain: C: PDB Molecule:matrix protein m2;
PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
|
| 32 | c1nyjB_ |
|
not modelled |
11.8 |
50 |
PDB header:viral protein
Chain: B: PDB Molecule:matrix protein m2;
PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
|
| 33 | c1nyjA_ |
|
not modelled |
11.8 |
50 |
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein m2;
PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
|
| 34 | c1nyjD_ |
|
not modelled |
11.8 |
50 |
PDB header:viral protein
Chain: D: PDB Molecule:matrix protein m2;
PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
|
| 35 | c2z7eB_ |
|
not modelled |
10.1 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:nifu-like protein;
PDBTitle: crystal structure of aquifex aeolicus iscu with bound [2fe-2 2s] cluster
|
| 36 | d1jhda2 |
|
not modelled |
9.7 |
39 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 37 | c2qq4A_ |
|
not modelled |
9.5 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:iron-sulfur cluster biosynthesis protein iscu;
PDBTitle: crystal structure of iron-sulfur cluster biosynthesis2 protein iscu (ttha1736) from thermus thermophilus hb8
|
| 38 | d1v47a2 |
|
not modelled |
8.8 |
28 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 39 | d1sddb1 |
|
not modelled |
8.3 |
25 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 40 | d1sisa_ |
|
not modelled |
8.2 |
25 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Short-chain scorpion toxins |
| 41 | c2d7cD_ |
|
not modelled |
7.6 |
31 |
PDB header:protein transport
Chain: D: PDB Molecule:rab11 family-interacting protein 3;
PDBTitle: crystal structure of human rab11 in complex with fip3 rab-2 binding domain
|
| 42 | c3k2gA_ |
|
not modelled |
7.5 |
22 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
| 43 | d1qqga2 |
|
not modelled |
7.2 |
50 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:Phosphotyrosine-binding domain (PTB) |
| 44 | d1chla_ |
|
not modelled |
7.1 |
33 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Short-chain scorpion toxins |
| 45 | c2ljcA_ |
|
not modelled |
7.0 |
50 |
PDB header:transport protein/inhibitor
Chain: A: PDB Molecule:m2 protein, bm2 protein chimera;
PDBTitle: structure of the influenza am2-bm2 chimeric channel bound to2 rimantadine
|
| 46 | c2kadC_ |
|
not modelled |
7.0 |
50 |
PDB header:membrane protein
Chain: C: PDB Molecule:transmembrane peptide of matrix protein 2;
PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
|
| 47 | c2kadB_ |
|
not modelled |
7.0 |
50 |
PDB header:membrane protein
Chain: B: PDB Molecule:transmembrane peptide of matrix protein 2;
PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
|
| 48 | c2kadA_ |
|
not modelled |
7.0 |
50 |
PDB header:membrane protein
Chain: A: PDB Molecule:transmembrane peptide of matrix protein 2;
PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
|
| 49 | c2kadD_ |
|
not modelled |
7.0 |
50 |
PDB header:membrane protein
Chain: D: PDB Molecule:transmembrane peptide of matrix protein 2;
PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
|
| 50 | d1xx6a1 |
|
not modelled |
7.0 |
29 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Type II thymidine kinase |
| 51 | c2jnhA_ |
|
not modelled |
7.0 |
54 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase cbl-b;
PDBTitle: solution structure of the uba domain from cbl-b
|
| 52 | d1p5ta_ |
|
not modelled |
6.7 |
40 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:Phosphotyrosine-binding domain (PTB) |
| 53 | c2vefB_ |
|
not modelled |
6.5 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 54 | c1jhdA_ |
|
not modelled |
6.4 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: crystal structure of bacterial atp sulfurylase from the2 riftia pachyptila symbiont
|
| 55 | d2arha1 |
|
not modelled |
6.4 |
40 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:Aq 1966-like |
| 56 | d1bm9a_ |
|
not modelled |
6.2 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Replication terminator protein (RTP) |
| 57 | c2p2uA_ |
|
not modelled |
5.9 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:host-nuclease inhibitor protein gam, putative;
PDBTitle: crystal structure of putative host-nuclease inhibitor2 protein gam from desulfovibrio vulgaris
|
| 58 | c2d9sA_ |
|
not modelled |
5.7 |
31 |
PDB header:ligase
Chain: A: PDB Molecule:cbl e3 ubiquitin protein ligase;
PDBTitle: solution structure of rsgi ruh-049, a uba domain from mouse2 cdna
|
| 59 | d1gota2 |
|
not modelled |
5.5 |
40 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:G proteins |
| 60 | c2dzaA_ |
|
not modelled |
5.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 61 | c2jayA_ |
|
not modelled |
5.3 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:proteasome;
PDBTitle: proteasome beta subunit prcb from mycobacterium2 tuberculosis
|
| 62 | c2do6A_ |
|
not modelled |
5.2 |
54 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase cbl-b;
PDBTitle: solution structure of rsgi ruh-065, a uba domain from human2 cdna
|
| 63 | c1ewrA_ |
|
not modelled |
5.2 |
43 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: crystal structure of taq muts
|