1 c1l5jB_
100.0
100
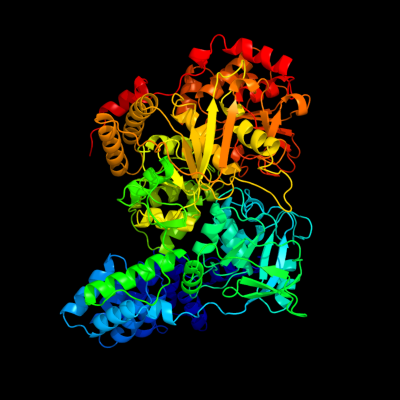
PDB header: lyaseChain: B: PDB Molecule: aconitate hydratase 2;PDBTitle: crystal structure of e. coli aconitase b.
2 d1l5ja3
100.0
100
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain3 d1acoa2
100.0
22
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain4 c5acnA_
100.0
22
PDB header: lyase(carbon-oxygen)Chain: A: PDB Molecule: aconitase;PDBTitle: structure of activated aconitase. formation of the (4fe-4s)2 cluster in the crystal
5 d2b3ya2
100.0
22
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain6 c2b3yB_
100.0
22
PDB header: lyaseChain: B: PDB Molecule: iron-responsive element binding protein 1;PDBTitle: structure of a monoclinic crystal form of human cytosolic aconitase2 (irp1)
7 d1l5ja1
100.0
100
Fold: alpha-alpha superhelixSuperfamily: Aconitase B, N-terminal domainFamily: Aconitase B, N-terminal domain8 d1l5ja2
100.0
100
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like9 d1v7la_
99.9
28
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like10 c2pkpA_
99.9
20
PDB header: lyaseChain: A: PDB Molecule: homoaconitase small subunit;PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
11 d2b3ya1
99.7
25
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like12 d1acoa1
99.7
22
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like13 c2hcuA_
99.6
18
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus2 mutans
14 c3q3wB_
99.6
16
PDB header: transferaseChain: B: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
15 c3h5jA_
99.5
20
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
16 c3t07D_
87.8
20
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
17 c2e28A_
72.5
22
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
18 d1kbla2
54.6
20
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain19 d1vbga2
53.4
15
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain20 d1h6za2
45.4
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain21 d1xdpa3
not modelled
42.0
11
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain22 d2fe0a1
not modelled
37.9
30
Fold: Smp-1-likeSuperfamily: Smp-1-likeFamily: Smp-1-like23 d2hi6a1
not modelled
35.7
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: AF0055-like24 d2gp4a1
not modelled
35.6
32
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like25 c2gp4A_
not modelled
34.3
30
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
26 d2z1ca1
not modelled
31.5
11
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like27 d2ot2a1
not modelled
27.0
17
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like28 c2gefA_
not modelled
24.9
28
PDB header: hydrolaseChain: A: PDB Molecule: protease vp4;PDBTitle: crystal structure of a novel viral protease with a2 serine/lysine catalytic dyad mechanism
29 d1zyma2
not modelled
23.0
14
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system30 c2fsdB_
not modelled
22.8
29
PDB header: viral proteinChain: B: PDB Molecule: putative baseplate protein;PDBTitle: a common fold for the receptor binding domains of2 lactococcal phages? the crystal structure of the head3 domain of phage bil170
31 d1kjna_
not modelled
22.3
16
Fold: Hypothetical protein MTH777 (MT0777)Superfamily: Hypothetical protein MTH777 (MT0777)Family: Hypothetical protein MTH777 (MT0777)32 c3d0fA_
not modelled
21.2
19
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
33 d1uw4a_
not modelled
21.2
18
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Smg-4/UPF334 c3pcoC_
not modelled
20.7
23
PDB header: ligaseChain: C: PDB Molecule: phenylalanyl-trna synthetase, alpha subunit;PDBTitle: crystal structure of e. coli phenylalanine-trna synthetase complexed2 with phenylalanine and amp
35 c3r74B_
not modelled
19.5
21
PDB header: lyase, biosynthetic proteinChain: B: PDB Molecule: anthranilate/para-aminobenzoate synthases component i;PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
36 c3d3rA_
not modelled
19.5
18
PDB header: chaperoneChain: A: PDB Molecule: hydrogenase assembly chaperone hypc/hupf;PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
37 c1dgsB_
not modelled
19.3
23
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
38 d2ivda2
not modelled
19.2
29
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase39 c1ni5A_
not modelled
18.9
9
PDB header: cell cycleChain: A: PDB Molecule: putative cell cycle protein mesj;PDBTitle: structure of the mesj pp-atpase from escherichia coli
40 d3d3ra1
not modelled
18.9
18
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like41 d1obra_
not modelled
18.9
22
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Carboxypeptidase T42 c2ebbA_
not modelled
18.5
19
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
43 c3k2gA_
not modelled
18.2
14
PDB header: resiniferatoxin binding proteinChain: A: PDB Molecule: resiniferatoxin-binding, phosphotriesterase-PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
44 d1r5ta_
not modelled
17.8
22
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase45 c2fgxA_
not modelled
17.7
40
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thioredoxin;PDBTitle: solution nmr structure of protein ne2328 from nitrosomonas2 europaea. northeast structural genomics consortium target3 net3.
46 c3l4gI_
not modelled
17.5
21
PDB header: ligaseChain: I: PDB Molecule: phenylalanyl-trna synthetase alpha chain;PDBTitle: crystal structure of homo sapiens cytoplasmic phenylalanyl-trna2 synthetase
47 d2pjua1
not modelled
17.3
19
Fold: Chelatase-likeSuperfamily: PrpR receptor domain-likeFamily: PrpR receptor domain-like48 c2xecD_
not modelled
17.1
9
PDB header: isomeraseChain: D: PDB Molecule: putative maleate isomerase;PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
49 c3b8fB_
not modelled
17.1
18
PDB header: hydrolaseChain: B: PDB Molecule: putative blasticidin s deaminase;PDBTitle: crystal structure of the cytidine deaminase from bacillus anthracis
50 d1elka_
not modelled
17.0
21
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain51 d1uwza_
not modelled
16.5
9
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase52 d2d30a1
not modelled
16.5
17
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase53 c3r2nC_
not modelled
16.3
12
PDB header: hydrolaseChain: C: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium leprae
54 c2yb1A_
not modelled
16.3
19
PDB header: hydrolaseChain: A: PDB Molecule: amidohydrolase;PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
55 c2kenA_
not modelled
15.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: solution nmr structure of the ob domain (residues 67-166)2 of mm0293 from methanosarcina mazei. northeast structural3 genomics consortium target mar214a.
56 c2duwA_
not modelled
15.8
21
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
57 c2q5cA_
not modelled
15.8
13
PDB header: transcriptionChain: A: PDB Molecule: ntrc family transcriptional regulator;PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
58 c3dcpB_
not modelled
15.5
30
PDB header: hydrolaseChain: B: PDB Molecule: histidinol-phosphatase;PDBTitle: crystal structure of the putative histidinol phosphatase2 hisk from listeria monocytogenes. northeast structural3 genomics consortium target lmr141.
59 c3bolB_
not modelled
15.3
23
PDB header: transferaseChain: B: PDB Molecule: 5-methyltetrahydrofolate s-homocysteinePDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
60 d2jnaa1
not modelled
15.3
13
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like61 c3h8aF_
not modelled
15.0
27
PDB header: lyase/protein bindingChain: F: PDB Molecule: rnase e;PDBTitle: crystal structure of e. coli enolase bound to its cognate rnase e2 recognition domain
62 c2jz2A_
not modelled
14.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ssl0352 protein;PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
63 c1pt1B_
not modelled
14.7
20
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
64 c2e21A_
not modelled
14.7
12
PDB header: ligaseChain: A: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
65 d1gg2g_
not modelled
14.6
30
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain66 d2gysa1
not modelled
14.6
31
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III67 d1w44a_
not modelled
14.4
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)68 c3h8aE_
not modelled
14.3
27
PDB header: lyase/protein bindingChain: E: PDB Molecule: rnase e;PDBTitle: crystal structure of e. coli enolase bound to its cognate rnase e2 recognition domain
69 c2owoA_
not modelled
14.1
23
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
70 d1seza2
not modelled
13.9
24
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase71 d2fr5a1
not modelled
13.8
9
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase72 d2cb5a_
not modelled
13.7
23
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Papain-like73 c1ytzI_
not modelled
13.6
25
PDB header: contractile proteinChain: I: PDB Molecule: troponin i;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 activated state
74 d2ga5a1
not modelled
13.6
29
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like75 c2fhdA_
not modelled
13.5
26
PDB header: cell cycleChain: A: PDB Molecule: dna repair protein rhp9/crb2;PDBTitle: crystal structure of crb2 tandem tudor domains
76 c3dmoD_
not modelled
13.5
8
PDB header: hydrolaseChain: D: PDB Molecule: cytidine deaminase;PDBTitle: 1.6 a crystal structure of cytidine deaminase from2 burkholderia pseudomallei
77 d1iuka_
not modelled
13.4
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain78 d1ppya_
not modelled
13.4
20
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC79 c2l33A_
not modelled
13.4
27
PDB header: transcription regulatorChain: A: PDB Molecule: interleukin enhancer-binding factor 3;PDBTitle: solution nmr structure of drbm 2 domain of interleukin enhancer-2 binding factor 3 from homo sapiens, northeast structural genomics3 consortium target hr4527e
80 d1jjca_
not modelled
13.3
20
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain81 c3imoC_
not modelled
13.3
26
PDB header: unknown functionChain: C: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass14
82 d2fz5a1
not modelled
13.3
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related83 c6r1rD_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: D: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441d mutant r1 protein from2 escherichia coli
84 c5r1rD_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: D: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441a mutant r1 protein from2 escherichia coli
85 c7r1rE_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: E: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441q mutant r1 protein from2 escherichia coli
86 c7r1rD_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: D: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441q mutant r1 protein from2 escherichia coli
87 c6r1rE_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: E: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441d mutant r1 protein from2 escherichia coli
88 c5r1rE_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: E: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441a mutant r1 protein from2 escherichia coli
89 c5r1rF_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: F: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441a mutant r1 protein from2 escherichia coli
90 c7r1rF_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: F: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441q mutant r1 protein from2 escherichia coli
91 c6r1rF_
not modelled
13.2
55
PDB header: complex (oxidoreductase/peptide)Chain: F: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase e441d mutant r1 protein from2 escherichia coli
92 c2xaxD_
not modelled
13.0
55
PDB header: oxidoreductaseChain: D: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y730no2y and y731a modified r12 subunit of e. coli
93 c2xakD_
not modelled
13.0
55
PDB header: oxidoreductaseChain: D: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y730no2y modified r1 subunit of e.2 coli
94 c2x1yF_
not modelled
13.0
55
PDB header: oxidoreductaseChain: F: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y731no2y modified r1 subunit of e.2 coli to 2.1 a resolution
95 c2x3sE_
not modelled
13.0
55
PDB header: oxidoreductaseChain: E: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y730no2y and c439a modified r12 subunit of e. coli
96 c2x33F_
not modelled
13.0
55
PDB header: oxidoreductaseChain: F: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y731no2y and y730f modified r12 subunit of e. coli
97 c2x3sF_
not modelled
13.0
55
PDB header: oxidoreductaseChain: F: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y730no2y and c439a modified r12 subunit of e. coli
98 c1r1rF_
not modelled
13.0
55
PDB header: oxidoreductaseChain: F: PDB Molecule: ribonucleotide reductase r2 protein;PDBTitle: ribonucleotide reductase r1 protein mutant y730f with a2 reduced active site from escherichia coli
99 c2x0zF_
not modelled
13.0
55
PDB header: oxidoreductaseChain: F: PDB Molecule: ribonucleoside-diphosphate reductase 1 subunitPDBTitle: ribonucleotide reductase y730no2y modified r1 subunit of e.2 coli