1 c3mk7K_
37.9
20
PDB header: oxidoreductaseChain: K: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit n;PDBTitle: the structure of cbb3 cytochrome oxidase
2 c3eh4A_
33.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase subunit 1;PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
3 c2k21A_
28.4
20
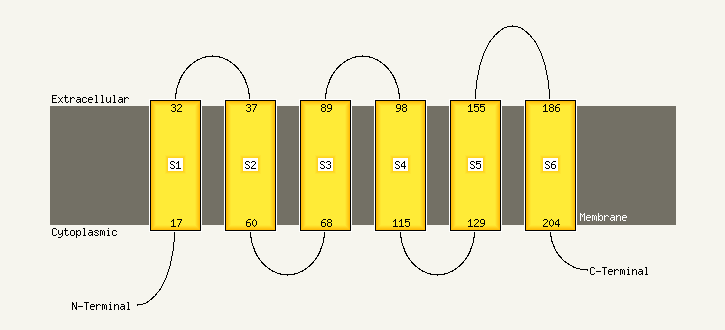
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
4 d1xmea1
27.6
17
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like5 d1pw4a_
26.2
16
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter6 d1pv7a_
25.9
12
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter7 d2ecba1
25.6
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain8 c2zmeA_
22.8
27
PDB header: protein transportChain: A: PDB Molecule: vacuolar-sorting protein snf8;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
9 c3dinD_
21.1
7
PDB header: membrane protein, protein transportChain: D: PDB Molecule: preprotein translocase subunit sece;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
10 c3pmiC_
19.3
18
PDB header: protein bindingChain: C: PDB Molecule: pwwp domain-containing protein mum1;PDBTitle: pwwp domain of human mutated melanoma-associated antigen 1
11 c2jp3A_
13.4
16
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
12 c1u5tA_
12.9
3
PDB header: transport proteinChain: A: PDB Molecule: appears to be functionally related to snf7;PDBTitle: structure of the escrt-ii endosomal trafficking complex
13 d1ha7b_
12.3
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins14 d1b8da_
12.0
18
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins15 c2ntxB_
10.9
36
PDB header: signaling proteinChain: B: PDB Molecule: emb16 d1u5ta1
8.4
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain17 d1cpca_
8.1
5
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins18 c3mlqD_
8.0
29
PDB header: transferase/transcriptionChain: D: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
19 d1phnb_
8.0
18
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins20 d1eyxb_
7.9
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins21 c3mlqB_
not modelled
7.8
29
PDB header: transferase/transcriptionChain: B: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
22 c2jo1A_
not modelled
7.6
9
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
23 d1a6qa1
not modelled
7.6
80
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain24 d1v54a_
not modelled
7.4
9
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like25 c1qfqB_
not modelled
7.3
43
PDB header: transcription/rnaChain: B: PDB Molecule: 36-mer n-terminal peptide of the n protein;PDBTitle: bacteriophage lambda n-protein-nutboxb-rna complex
26 c2c7jB_
not modelled
7.2
18
PDB header: electron transportChain: B: PDB Molecule: phycoerythrocyanin beta chain;PDBTitle: phycoerythrocyanin from mastigocladus laminosus, 295 k,2 3.0 a
27 d1ffta_
not modelled
7.0
9
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like28 c1fftF_
not modelled
7.0
9
PDB header: oxidoreductaseChain: F: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
29 c2e19A_
not modelled
6.8
6
PDB header: transcriptionChain: A: PDB Molecule: transcription factor 8;PDBTitle: solution structure of the homeobox domain from human nil-2-2 a zinc finger protein, transcription factor 8
30 d1cpcb_
not modelled
6.8
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins31 c3mp7A_
not modelled
6.7
50
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase subunit secy;PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
32 c2kncA_
not modelled
6.7
8
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
33 d1hynp_
not modelled
6.6
26
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain34 c2rddB_
not modelled
6.6
14
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
35 d1liab_
not modelled
6.5
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins36 c1hynQ_
not modelled
6.5
16
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
37 d1kn1a_
not modelled
6.4
21
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins38 d1ar1a_
not modelled
6.4
2
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like39 d1xmec1
not modelled
6.3
53
Fold: Single transmembrane helixSuperfamily: Bacterial ba3 type cytochrome c oxidase subunit IIaFamily: Bacterial ba3 type cytochrome c oxidase subunit IIa40 d1f99b_
not modelled
6.2
20
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins41 c2bbjB_
not modelled
6.1
17
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
42 c3smaD_
not modelled
6.0
41
PDB header: transferaseChain: D: PDB Molecule: frbf;PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
43 d1vp8a_
not modelled
5.9
25
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like44 c2vmlD_
not modelled
5.8
13
PDB header: photosynthesisChain: D: PDB Molecule: phycocyanin beta chain;PDBTitle: the monoclinic structure of phycocyanin from gloeobacter2 violaceus
45 c3hwpA_
not modelled
5.8
23
PDB header: hydrolaseChain: A: PDB Molecule: phlg;PDBTitle: crystal structure and computational analyses provide insights into the2 catalytic mechanism of 2, 4-diacetylphloroglucinol hydrolase phlg3 from pseudomonas fluorescens
46 c2da7A_
not modelled
5.8
9
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger homeobox protein 1b;PDBTitle: solution structure of the homeobox domain of zinc finger2 homeobox protein 1b (smad interacting protein 1)
47 c3e4fB_
not modelled
5.8
41
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside n3-acetyltransferase;PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
48 c1cw5A_
not modelled
5.7
29
PDB header: toxinChain: A: PDB Molecule: type iia bacteriocin carnobacteriocin b2;PDBTitle: solution structure of carnobacteriocin b2
49 d2h8pc1
not modelled
5.7
11
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels50 d1mtyg_
not modelled
5.7
15
Fold: Open three-helical up-and-down bundleSuperfamily: Methane monooxygenase hydrolase, gamma subunitFamily: Methane monooxygenase hydrolase, gamma subunit51 c1jb0K_
not modelled
5.7
11
PDB header: photosynthesisChain: K: PDB Molecule: photosystem 1 reaction centre subunit x;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
52 d1jb0k_
not modelled
5.7
11
Fold: Photosystem I reaction center subunit X, PsaKSuperfamily: Photosystem I reaction center subunit X, PsaKFamily: Photosystem I reaction center subunit X, PsaK53 c3gitA_
not modelled
5.7
10
PDB header: transferaseChain: A: PDB Molecule: carbon monoxide dehydrogenase/acetyl-coa synthase subunitPDBTitle: crystal structure of a truncated acetyl-coa synthase
54 d1unld_
not modelled
5.6
29
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin55 c1q90L_
not modelled
5.6
38
PDB header: photosynthesisChain: L: PDB Molecule: cytochrome b6f complex subunit petl;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
56 d1q90l_
not modelled
5.6
38
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex57 d1mhyg_
not modelled
5.6
10
Fold: Open three-helical up-and-down bundleSuperfamily: Methane monooxygenase hydrolase, gamma subunitFamily: Methane monooxygenase hydrolase, gamma subunit58 c2ka2A_
not modelled
5.4
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
59 c2ka1B_
not modelled
5.4
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
60 c2ka1A_
not modelled
5.4
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
61 c2ka2B_
not modelled
5.4
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
62 d1ha7a_
not modelled
5.4
14
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins63 d1jboa_
not modelled
5.4
10
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins64 c1m56G_
not modelled
5.3
5
PDB header: oxidoreductaseChain: G: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
65 c1peiA_
not modelled
5.3
50
PDB header: nucleotidyltransferaseChain: A: PDB Molecule: pepc22;PDBTitle: nmr structure of the membrane-binding domain of ctp2 phosphocholine cytidylyltransferase, 10 structures
66 d1xg0c_
not modelled
5.2
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins67 c2kluA_
not modelled
5.1
23
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
68 c2diiA_
not modelled
5.1
23
PDB header: transcriptionChain: A: PDB Molecule: tfiih basal transcription factor complex p62PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
69 c1ifpA_
not modelled
5.1
32
PDB header: virusChain: A: PDB Molecule: major coat protein assembly;PDBTitle: inovirus (filamentous bacteriophage) strain pf3 major coat2 protein assembly
70 d1sana_
not modelled
5.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain71 d1f99a_
not modelled
5.1
7
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins