| 1 | c2x0kB_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribf;
PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
|
| 2 | c3op1A_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:macrolide-efflux protein;
PDBTitle: crystal structure of macrolide-efflux protein sp_1110 from2 streptococcus pneumoniae
|
| 3 | c1t6zB_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin kinase/fmn adenylyltransferase;
PDBTitle: crystal structure of riboflavin bound tm379
|
| 4 | d1nb9a_
|
|
 |
100.0 |
27 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 5 | d1n08a_
|
|
 |
100.0 |
30 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 6 | c3bnwA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin kinase, putative;
PDBTitle: crystal structure of riboflavin kinase from trypanosoma brucei
|
| 7 | d1mrza1
|
|
 |
100.0 |
43 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 8 | d1mrza2
|
|
 |
100.0 |
27 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 9 | c3guzB_
|
|
 |
99.9 |
23 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
|
| 10 | c3glvB_
|
|
 |
99.8 |
19 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:lipopolysaccharide core biosynthesis protein;
PDBTitle: crystal structure of the lipopolysaccharide core biosynthesis protein2 from thermoplasma volcanium gss1
|
| 11 | d1coza_
|
|
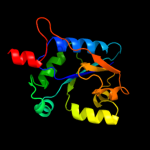 |
99.7 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Cytidylyltransferase |
| 12 | c1lw7A_
|
|
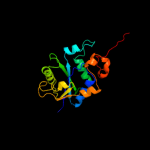 |
99.7 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:transcriptional regulator nadr;
PDBTitle: nadr protein from haemophilus influenzae
|
| 13 | c3uk2B_
|
|
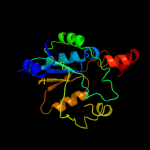 |
99.7 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
|
| 14 | c2ejcA_
|
|
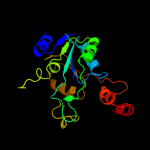 |
99.6 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:pantoate--beta-alanine ligase;
PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
|
| 15 | d1v8fa_
|
|
 |
99.6 |
23 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
| 16 | d1ihoa_
|
|
 |
99.6 |
24 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
| 17 | c3h05A_
|
|
 |
99.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vpa0413;
PDBTitle: the crystal structure of a putative nicotinate-nucleotide2 adenylyltransferase from vibrio parahaemolyticus
|
| 18 | d1lw7a1
|
|
 |
99.6 |
10 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 19 | c3n8hA_
|
|
 |
99.6 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
|
| 20 | c3e27B_
|
|
 |
99.5 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:nicotinate (nicotinamide) nucleotide
PDBTitle: nicotinic acid mononucleotide (namn) adenylyltransferase2 from bacillus anthracis: product complex
|
| 21 | c3elbA_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:ethanolamine-phosphate cytidylyltransferase;
PDBTitle: human ctp: phosphoethanolamine cytidylyltransferase in complex with2 cmp
|
| 22 | c3ag5A_ |
|
not modelled |
99.5 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantothenate synthetase from staphylococcus2 aureus
|
| 23 | c2h29A_ |
|
not modelled |
99.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from staphylococcus aureus: product3 bound form 1
|
| 24 | c3innB_ |
|
not modelled |
99.5 |
25 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine-ligase in complex2 with atp at low occupancy at 2.1 a resolution
|
| 25 | c2b7lD_ |
|
not modelled |
99.5 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:glycerol-3-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ctp:glycerol-3-phosphate2 cytidylyltransferase from staphylococcus aureus
|
| 26 | c3nv7A_ |
|
not modelled |
99.5 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of h.pylori phosphopantetheine adenylyltransferase2 mutant i4v/n76y
|
| 27 | c3mxtA_ |
|
not modelled |
99.4 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
|
| 28 | d1f9aa_ |
|
not modelled |
99.4 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 29 | d1qjca_ |
|
not modelled |
99.4 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 30 | d2a84a1 |
|
not modelled |
99.4 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
| 31 | d1kr2a_ |
|
not modelled |
99.4 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 32 | d1nuua_ |
|
not modelled |
99.4 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 33 | d1tfua_ |
|
not modelled |
99.3 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 34 | d1o6ba_ |
|
not modelled |
99.3 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 35 | d1ej2a_ |
|
not modelled |
99.3 |
23 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 36 | c3ikzA_ |
|
not modelled |
99.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of phosphopantetheine adenylyltransferase from2 burkholderia pseudomallei
|
| 37 | c3hl4B_ |
|
not modelled |
99.2 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:choline-phosphate cytidylyltransferase a;
PDBTitle: crystal structure of a mammalian ctp:phosphocholine2 cytidylyltransferase with cdp-choline
|
| 38 | d1vlha_ |
|
not modelled |
99.1 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 39 | c3nd5D_ |
|
not modelled |
98.9 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of phosphopantetheine adenylyltransferase (ppat)2 from enterococcus faecalis
|
| 40 | d1od6a_ |
|
not modelled |
98.9 |
22 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 41 | c2r5wA_ |
|
not modelled |
98.9 |
14 |
PDB header:hydrolase, transferase
Chain: A: PDB Molecule:nicotinamide-nucleotide adenylyltransferase;
PDBTitle: crystal structure of a bifunctional nmn2 adenylyltransferase/adp ribose pyrophosphatase from3 francisella tularensis
|
| 42 | c2qjoB_ |
|
not modelled |
98.8 |
16 |
PDB header:transferase, hydrolase
Chain: B: PDB Molecule:bifunctional nmn adenylyltransferase/nudix hydrolase;
PDBTitle: crystal structure of a bifunctional nmn adenylyltransferase/adp ribose2 pyrophosphatase (nadm) complexed with adprp and nad from3 synechocystis sp.
|
| 43 | c1yunB_ |
|
not modelled |
98.8 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from pseudomonas aeruginosa
|
| 44 | c3f3mA_ |
|
not modelled |
98.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: six crystal structures of two phosphopantetheine2 adenylyltransferases reveal an alternative ligand binding3 mode and an associated structural change
|
| 45 | d1kama_ |
|
not modelled |
98.7 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 46 | d1k4ma_ |
|
not modelled |
98.6 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 47 | c3gmiA_ |
|
not modelled |
98.6 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0348 protein mj0951;
PDBTitle: crystal structure of a protein of unknown function from2 methanocaldococcus jannaschii
|
| 48 | c3do8B_ |
|
not modelled |
98.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: the crystal structure of the protein with unknown function2 from archaeoglobus fulgidus
|
| 49 | d1jhda2 |
|
not modelled |
97.9 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 50 | c2qjfB_ |
|
not modelled |
97.4 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional 3'-phosphoadenosine 5'-
PDBTitle: crystal structure of atp-sulfurylase domain of human paps2 synthetase 1
|
| 51 | c1xnjB_ |
|
not modelled |
97.3 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional 3'-phosphoadenosine 5'-phosphosulfate
PDBTitle: aps complex of human paps synthetase 1
|
| 52 | d1x6va2 |
|
not modelled |
97.3 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 53 | d1g8fa2 |
|
not modelled |
96.7 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 54 | c1r6xA_ |
|
not modelled |
96.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:atp:sulfate adenylyltransferase;
PDBTitle: the crystal structure of a truncated form of yeast atp2 sulfurylase, lacking the c-terminal aps kinase-like domain,3 in complex with sulfate
|
| 55 | c1jhdA_ |
|
not modelled |
96.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: crystal structure of bacterial atp sulfurylase from the2 riftia pachyptila symbiont
|
| 56 | d1v47a2 |
|
not modelled |
96.3 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 57 | d1m8pa2 |
|
not modelled |
96.1 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 58 | c1g8gB_ |
|
not modelled |
96.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: atp sulfurylase from s. cerevisiae: the binary product complex with2 aps
|
| 59 | c1m8pB_ |
|
not modelled |
95.5 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: crystal structure of p. chrysogenum atp sulfurylase in the t-state
|
| 60 | c3cr8C_ |
|
not modelled |
94.5 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:sulfate adenylyltranferase, adenylylsulfate
PDBTitle: hexameric aps kinase from thiobacillus denitrificans
|
| 61 | c2gksB_ |
|
not modelled |
94.4 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional sat/aps kinase;
PDBTitle: crystal structure of the bi-functional atp sulfurylase-aps kinase from2 aquifex aeolicus, a chemolithotrophic thermophile
|
| 62 | c1v47B_ |
|
not modelled |
92.1 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:atp sulfurylase;
PDBTitle: crystal structure of atp sulfurylase from thermus2 thermophillus hb8 in complex with aps
|
| 63 | c3ih5A_ |
|
not modelled |
79.9 |
20 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: crystal structure of electron transfer flavoprotein alpha-2 subunit from bacteroides thetaiotaomicron
|
| 64 | c1dkrB_ |
|
not modelled |
68.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 65 | d2g2ca1 |
|
not modelled |
64.2 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 66 | c3bdkB_ |
|
not modelled |
60.2 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 67 | c3lx4B_ |
|
not modelled |
57.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fe-hydrogenase;
PDBTitle: stepwise [fefe]-hydrogenase h-cluster assembly revealed in the2 structure of hyda(deltaefg)
|
| 68 | c3dahB_ |
|
not modelled |
55.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
| 69 | d1jvna1 |
|
not modelled |
55.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 70 | d2nqra3 |
|
not modelled |
53.9 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 71 | c3rfqC_ |
|
not modelled |
53.3 |
22 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
| 72 | c2is8A_ |
|
not modelled |
50.2 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 73 | c1cvrA_ |
|
not modelled |
50.0 |
15 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:gingipain r;
PDBTitle: crystal structure of the arg specific cysteine proteinase gingipain r2 (rgpb)
|
| 74 | d2h85a2 |
|
not modelled |
49.5 |
15 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 75 | c3jxeB_ |
|
not modelled |
46.2 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of pyrococcus horikoshii tryptophanyl-trna2 synthetase in complex with trpamp
|
| 76 | c2ip1A_ |
|
not modelled |
46.1 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure analysis of s. cerevisiae tryptophanyl trna2 synthetase
|
| 77 | c3eb2A_ |
|
not modelled |
44.8 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 78 | c3kbqA_ |
|
not modelled |
44.4 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
| 79 | c1uz5A_ |
|
not modelled |
42.6 |
17 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
| 80 | c3focB_ |
|
not modelled |
41.6 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from giardia lamblia
|
| 81 | c2fu3A_ |
|
not modelled |
40.9 |
21 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
| 82 | d2ozka2 |
|
not modelled |
40.5 |
23 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 83 | c3hv0A_ |
|
not modelled |
40.2 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from cryptosporidium parvum
|
| 84 | d2choa2 |
|
not modelled |
40.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:alpha-D-glucuronidase/Hyaluronidase catalytic domain |
| 85 | c1jvnB_ |
|
not modelled |
39.6 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 86 | d1xxxa1 |
|
not modelled |
39.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 87 | d1np7a2 |
|
not modelled |
39.0 |
11 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 88 | c2hqxB_ |
|
not modelled |
38.4 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:p100 co-activator tudor domain;
PDBTitle: crystal structure of human p100 tudor domain conserved2 region
|
| 89 | d2hqxa1 |
|
not modelled |
38.4 |
11 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
| 90 | c1gcyA_ |
|
not modelled |
37.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucan 1,4-alpha-maltotetrahydrolase;
PDBTitle: high resolution crystal structure of maltotetraose-forming2 exo-amylase
|
| 91 | c2r8wB_ |
|
not modelled |
37.4 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 92 | d1y5ea1 |
|
not modelled |
36.8 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 93 | d1xi8a3 |
|
not modelled |
36.7 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 94 | d1mkza_ |
|
not modelled |
35.0 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 95 | c2zskA_ |
|
not modelled |
34.8 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
| 96 | d2gtia2 |
|
not modelled |
34.7 |
21 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 97 | c2g4rB_ |
|
not modelled |
34.5 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis mog protein;
PDBTitle: anomalous substructure of moga
|
| 98 | d1o5ka_ |
|
not modelled |
34.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 99 | c2eqkA_ |
|
not modelled |
34.0 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:tudor domain-containing protein 4;
PDBTitle: solution structure of the tudor domain of tudor domain-2 containing protein 4
|
| 100 | c2ya0A_ |
|
not modelled |
33.6 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
|
| 101 | c2xrzA_ |
|
not modelled |
33.6 |
17 |
PDB header:lyase/dna
Chain: A: PDB Molecule:deoxyribodipyrimidine photolyase;
PDBTitle: x-ray structure of archaeal class ii cpd photolyase from2 methanosarcina mazei in complex with intact cpd-lesion
|
| 102 | d1uz5a3 |
|
not modelled |
33.3 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 103 | d1owla2 |
|
not modelled |
33.1 |
15 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 104 | c2el7A_ |
|
not modelled |
32.3 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from thermus2 thermophilus
|
| 105 | d3clsd1 |
|
not modelled |
31.9 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 106 | c2rfgB_ |
|
not modelled |
31.6 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 107 | c2qlcC_ |
|
not modelled |
31.4 |
12 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna repair protein radc homolog;
PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
|
| 108 | d1hf2a2 |
|
not modelled |
31.3 |
20 |
Fold:Cell-division inhibitor MinC, N-terminal domain
Superfamily:Cell-division inhibitor MinC, N-terminal domain
Family:Cell-division inhibitor MinC, N-terminal domain |
| 109 | c2nqqA_ |
|
not modelled |
31.3 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
| 110 | d1lwha2 |
|
not modelled |
31.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 111 | d1wu2a3 |
|
not modelled |
31.2 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 112 | d2f7wa1 |
|
not modelled |
30.4 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 113 | c3i05B_ |
|
not modelled |
30.1 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from trypanosoma brucei
|
| 114 | c1ehaA_ |
|
not modelled |
29.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyltrehalose trehalohydrolase;
PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase2 from sulfolobus solfataricus
|
| 115 | d1r6ta2 |
|
not modelled |
29.6 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 116 | c3cvyA_ |
|
not modelled |
29.6 |
12 |
PDB header:lyase/dna
Chain: A: PDB Molecule:re11660p;
PDBTitle: drosophila melanogaster (6-4) photolyase bound to repaired2 ds dna
|
| 117 | c1wu2B_ |
|
not modelled |
29.5 |
15 |
PDB header:structural genomics,biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis moea protein;
PDBTitle: crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikoshii ot3
|
| 118 | c3zt5D_ |
|
not modelled |
29.5 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative glucanohydrolase pep1a;
PDBTitle: glge isoform 1 from streptomyces coelicolor with maltose2 bound
|
| 119 | c2gtiA_ |
|
not modelled |
29.4 |
16 |
PDB header:viral protein
Chain: A: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: mutation of mhv coronavirus non-structural protein nsp15 (f307l)
|
| 120 | c2by0A_ |
|
not modelled |
29.2 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:maltooligosyltrehalose trehalohydrolase;
PDBTitle: is radiation damage dependent on the dose-rate used during2 macromolecular crystallography data collection
|