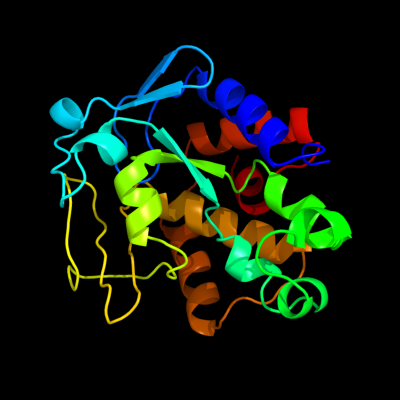
1 d1ouoa_
100.0
63
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Endonuclease I2 c2qgpA_
93.5
32
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
3 d2ebfx3
29.5
47
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: PMT C-terminal domain like4 c2e72A_
29.2
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pogo transposable element with znf domain;PDBTitle: solution structure of the zinc finger domain of human2 kiaa0461
5 c3c4rC_
23.4
24
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
6 d2ebfx1
20.9
12
Fold: PMT central region-likeSuperfamily: PMT central region-likeFamily: PMT central region-like7 c2lmdA_
18.9
22
PDB header: transcriptionChain: A: PDB Molecule: prospero homeobox protein 1;PDBTitle: minimal constraints solution nmr structure of prospero homeobox2 protein 1 from homo sapiens, northeast structural genomics consortium3 target hr4660b
8 d1mija_
15.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain9 c3gqeA_
15.4
16
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 3;PDBTitle: crystal structure of macro domain of venezuelan equine encephalitis2 virus
10 d2evra1
14.7
38
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: Spr N-terminal domain-like11 c1tr8A_
13.7
17
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
12 c2ec5B_
11.2
12
PDB header: toxinChain: B: PDB Molecule: dermonecrotic toxin;PDBTitle: crystal structures reveal a thiol-protease like catalytic triad in the2 c-terminal region of pasteurella multocida toxin
13 d1em8b_
11.1
21
Fold: DNA polymerase III psi subunitSuperfamily: DNA polymerase III psi subunitFamily: DNA polymerase III psi subunit14 c1m8oB_
10.6
26
PDB header: membrane proteinChain: B: PDB Molecule: platele integrin beta3 subunit: cytoplasmicPDBTitle: platelet integrin alfaiib-beta3 cytoplasmic domain
15 c3gpqA_
10.6
21
PDB header: viral protein/rnaChain: A: PDB Molecule: non-structural protein 3;PDBTitle: crystal structure of macro domain of chikungunya virus in complex with2 rna
16 d1yc611
9.7
28
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP17 c2bpbB_
9.6
10
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
18 c3g9wC_
9.1
20
PDB header: cell adhesionChain: C: PDB Molecule: integrin beta-1d;PDBTitle: crystal structure of talin2 f2-f3 in complex with the integrin beta1d2 cytoplasmic tail
19 d2bcgg3
8.4
42
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GDI-like20 c3t4aG_
8.0
33
PDB header: immune systemChain: G: PDB Molecule: fibrinogen-binding protein;PDBTitle: structure of a truncated form of staphylococcal complement inhibitor b2 bound to human c3c at 3.4 angstrom resolution
21 d1js9c_
not modelled
7.8
28
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP22 d1i5za1
not modelled
7.8
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like23 d1qbaa2
not modelled
7.7
18
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Bacterial chitobiase, n-terminal domain24 d2dk1a1
not modelled
7.6
18
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain25 c2a45L_
not modelled
7.5
25
PDB header: hydrolase/hydrolase inhibitorChain: L: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
26 c3gnzP_
not modelled
7.5
50
PDB header: toxinChain: P: PDB Molecule: 25 kda protein elicitor;PDBTitle: toxin fold for microbial attack and plant defense
27 c3bunB_
not modelled
7.3
24
PDB header: ligase/signaling proteinChain: B: PDB Molecule: e3 ubiquitin-protein ligase cbl;PDBTitle: crystal structure of c-cbl-tkb domain complexed with its2 binding motif in sprouty4
28 c2cblA_
not modelled
7.0
24
PDB header: complex (proto-oncogene/peptide)Chain: A: PDB Molecule: proto-oncogene cbl;PDBTitle: n-terminal domain of cbl in complex with its binding site2 on zap-70
29 d1yy9a4
not modelled
6.9
30
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Growth factor receptor domainFamily: Growth factor receptor domain30 c2qffA_
not modelled
6.8
27
PDB header: hydrolase inhibitorChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of staphylococcal complement inhibitor
31 d1cwpa_
not modelled
6.7
56
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP32 c3st1A_
not modelled
6.3
40
PDB header: toxinChain: A: PDB Molecule: necrosis-and ethylene-inducing protein;PDBTitle: crystal structure of necrosis and ethylene inducing protein 2 from the2 causal agent of cocoa's witches broom disease
33 c3nzqB_
not modelled
6.2
32
PDB header: lyaseChain: B: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
34 c2kncB_
not modelled
6.1
27
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
35 c2y69S_
not modelled
6.0
43
PDB header: electron transportChain: S: PDB Molecule: cytochrome c oxidase subunit 5b;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
36 d1d5ta2
not modelled
6.0
35
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GDI-like37 d3buxb2
not modelled
6.0
17
Fold: N-cbl likeSuperfamily: N-terminal domain of cbl (N-cbl)Family: N-terminal domain of cbl (N-cbl)38 d2gaua1
not modelled
5.9
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like39 c2yqfA_
not modelled
5.9
23
PDB header: protein bindingChain: A: PDB Molecule: ankyrin-1;PDBTitle: solution structure of the death domain of ankyrin-1
40 d1cwva5
not modelled
5.8
22
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Invasin/intimin cell-adhesion fragment, C-terminal domain41 c1cwpB_
not modelled
5.7
56
PDB header: virus/rnaChain: B: PDB Molecule: coat protein;PDBTitle: structures of the native and swollen forms of cowpea2 chlorotic mottle virus determined by x-ray crystallography3 and cryo-electron microscopy
42 c1hp9A_
not modelled
5.7
60
PDB header: toxinChain: A: PDB Molecule: kappa-hefutoxin 1;PDBTitle: kappa-hefutoxins: a novel class of potassium channel toxins2 from scorpion venom
43 d1pbya1
not modelled
5.3
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 244 c3rnvA_
not modelled
5.2
13
PDB header: hydrolaseChain: A: PDB Molecule: helper component proteinase;PDBTitle: structure of the autocatalytic cysteine protease domain of potyvirus2 helper-component proteinase
45 c2q4pA_
not modelled
5.0
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein rs21-c6;PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898