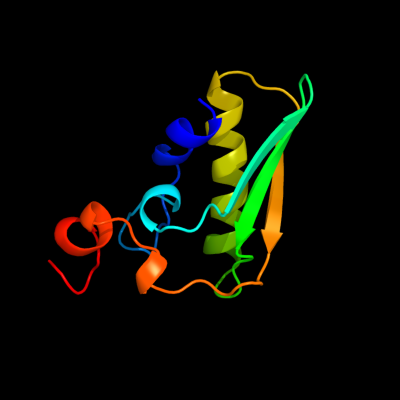
1 c3lnoA_
100.0
28
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
2 d1uwda_
100.0
27
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like3 d2cu6a1
99.9
30
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like4 c2z51A_
94.3
17
PDB header: metal transportChain: A: PDB Molecule: nifu-like protein 2, chloroplast;PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
5 d1xhja_
94.1
21
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like6 c2jnvA_
94.1
19
PDB header: metal transportChain: A: PDB Molecule: nifu-like protein 1, chloroplast;PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
7 d1veha_
89.3
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like8 d1th5a1
87.8
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like9 c3cw2M_
78.7
36
PDB header: translationChain: M: PDB Molecule: translation initiation factor 2 subunit beta;PDBTitle: crystal structure of the intact archaeal translation2 initiation factor 2 from sulfolobus solfataricus .
10 c3a44D_
74.0
27
PDB header: metal binding proteinChain: D: PDB Molecule: hydrogenase nickel incorporation protein hypa;PDBTitle: crystal structure of hypa in the dimeric form
11 c1neeA_
73.0
32
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 betaPDBTitle: structure of archaeal translation factor aif2beta from2 methanobacterium thermoautrophicum
12 d1k81a_
72.0
36
Fold: Zinc-binding domain of translation initiation factor 2 betaSuperfamily: Zinc-binding domain of translation initiation factor 2 betaFamily: Zinc-binding domain of translation initiation factor 2 beta13 d1dl6a_
70.4
19
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain14 c2kdxA_
69.6
21
PDB header: metal-binding proteinChain: A: PDB Molecule: hydrogenase/urease nickel incorporation proteinPDBTitle: solution structure of hypa protein
15 c2aklA_
65.1
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
16 d1wiia_
61.1
19
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Putative zinc binding domain17 d1pfta_
59.6
26
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain18 d1nuia2
58.4
29
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: DNA primase zinc finger19 d3bypa1
57.8
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like20 c2dcuB_
57.1
50
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 beta subunit;PDBTitle: crystal structure of translation initiation factor aif2betagamma2 heterodimer with gdp
21 d2akla2
not modelled
55.2
38
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: PhnA zinc-binding domain22 d1hk8a_
not modelled
52.0
21
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit23 c1hk8A_
not modelled
52.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
24 c2e9hA_
not modelled
50.3
43
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: solution structure of the eif-5_eif-2b domain from human2 eukaryotic translation initiation factor 5
25 d1lv3a_
not modelled
49.9
32
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Hypothetical zinc finger protein YacG26 c3lpeF_
not modelled
44.4
40
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
27 c2xigA_
not modelled
40.5
12
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
28 c3qxbB_
not modelled
39.0
18
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
29 c1x31D_
not modelled
38.6
36
PDB header: oxidoreductaseChain: D: PDB Molecule: sarcosine oxidase delta subunit;PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
30 c1qysA_
not modelled
37.9
13
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
31 c1eucB_
not modelled
37.4
15
PDB header: ligaseChain: B: PDB Molecule: succinyl-coa synthetase, beta chain;PDBTitle: crystal structure of dephosphorylated pig heart, gtp-2 specific succinyl-coa synthetase
32 c2kt2A_
not modelled
37.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
33 c2elpA_
not modelled
35.3
18
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
34 c2opfA_
not modelled
34.4
30
PDB header: hydrolase/dnaChain: A: PDB Molecule: endonuclease viii;PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
35 d1atga_
not modelled
34.3
24
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like36 c3zyqA_
not modelled
33.7
28
PDB header: signalingChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
37 d1qupa2
not modelled
33.6
9
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain38 c1k82D_
not modelled
33.3
20
PDB header: hydrolase/dnaChain: D: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
39 d2fhpa1
not modelled
33.2
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like40 d2gnra1
not modelled
33.0
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: SSO2064-like41 c2jvfA_
not modelled
32.7
17
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
42 d1j26a_
not modelled
32.0
23
Fold: dsRBD-likeSuperfamily: Peptidyl-tRNA hydrolase domain-likeFamily: Peptidyl-tRNA hydrolase domain43 d1pfva3
not modelled
31.6
40
Fold: Rubredoxin-likeSuperfamily: Methionyl-tRNA synthetase (MetRS), Zn-domainFamily: Methionyl-tRNA synthetase (MetRS), Zn-domain44 d2dsxa1
not modelled
31.4
14
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin45 c3mv2A_
not modelled
30.0
17
PDB header: protein transportChain: A: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of a-cop in complex with e-cop
46 d1iloa_
not modelled
28.0
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase47 c1nnjA_
not modelled
27.8
28
PDB header: hydrolaseChain: A: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna
48 c2rrlA_
not modelled
27.3
16
PDB header: protein transportChain: A: PDB Molecule: flagellar hook-length control protein;PDBTitle: solution structure of the c-terminal domain of the flik
49 c2fu4B_
not modelled
27.2
23
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
50 d2r7ca2
not modelled
27.1
71
Fold: Rotavirus NSP2 fragment, N-terminal domainSuperfamily: Rotavirus NSP2 fragment, N-terminal domainFamily: Rotavirus NSP2 fragment, N-terminal domain51 c2k2dA_
not modelled
26.8
31
PDB header: metal binding proteinChain: A: PDB Molecule: ring finger and chy zinc finger domain-PDBTitle: solution nmr structure of c-terminal domain of human pirh2.2 northeast structural genomics consortium (nesg) target ht2c
52 c2o03A_
not modelled
26.7
23
PDB header: gene regulationChain: A: PDB Molecule: probable zinc uptake regulation protein furb;PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
53 d2fiya1
not modelled
26.3
17
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like54 c2csuB_
not modelled
26.0
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 457aa long hypothetical protein;PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
55 d4rxna_
not modelled
25.5
15
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin56 c2hk1D_
not modelled
25.4
18
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
57 d1mzba_
not modelled
25.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like58 c2x5cB_
not modelled
24.4
33
PDB header: viral proteinChain: B: PDB Molecule: hypothetical protein orf131;PDBTitle: crystal structure of hypothetical protein orf131 from2 pyrobaculum spherical virus
59 c3jvfC_
not modelled
24.4
18
PDB header: signaling protein / cytokineChain: C: PDB Molecule: interleukin-17 receptor a;PDBTitle: crystal structure of an interleukin-17 receptor complex
60 c3eyyA_
not modelled
24.4
17
PDB header: transportChain: A: PDB Molecule: putative iron uptake regulatory protein;PDBTitle: structural basis for the specialization of nur, a nickel-2 specific fur homologue, in metal sensing and dna3 recognition
61 c2l7xA_
not modelled
24.2
60
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: crimean congo hemorrhagic fever gn zinc finger
62 c2w57A_
not modelled
24.0
26
PDB header: metal transportChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the vibrio cholerae ferric uptake2 regulator (fur) reveals structural rearrangement of the3 dna-binding domains
63 d1r7ha_
not modelled
23.9
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase64 c2nu9E_
not modelled
23.9
15
PDB header: ligaseChain: E: PDB Molecule: succinyl-coa synthetase beta chain;PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
65 d1xzoa1
not modelled
23.2
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like66 c2kn9A_
not modelled
23.2
13
PDB header: electron transportChain: A: PDB Molecule: rubredoxin;PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
67 d1dx8a_
not modelled
23.0
25
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin68 d1h7va_
not modelled
22.8
26
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin69 d1tz9a_
not modelled
22.7
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: UxuA-like70 c1s24A_
not modelled
22.6
17
PDB header: electron transportChain: A: PDB Molecule: rubredoxin 2;PDBTitle: rubredoxin domain ii from pseudomonas oleovorans
71 d1s24a_
not modelled
22.6
17
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin72 c2f5qA_
not modelled
22.6
24
PDB header: hydrolase/dnaChain: A: PDB Molecule: formamidopyrimidine-dna glycosidase;PDBTitle: catalytically inactive (e3q) mutm crosslinked to oxog:c2 containing dna cc2
73 c3mkrB_
not modelled
22.5
29
PDB header: transport proteinChain: B: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of yeast alpha/epsilon-cop subcomplex of the copi2 vesicular coat
74 c2pptA_
not modelled
22.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-2;PDBTitle: crystal structure of thioredoxin-2
75 d1iroa_
not modelled
21.9
15
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin76 d2fu5a1
not modelled
21.8
50
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss477 c3ktcB_
not modelled
21.7
20
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
78 d1ee8a3
not modelled
21.6
24
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins79 d2ct1a1
not modelled
21.6
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H280 d1brfa_
not modelled
21.4
17
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin81 d1cmca_
not modelled
21.3
44
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Met repressor, MetJ (MetR)82 d1qcva_
not modelled
21.2
10
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin83 d1rb9a_
not modelled
21.0
14
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin84 d1w4ha1
not modelled
20.7
28
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex85 d1vbka2
not modelled
20.5
20
Fold: THUMP domainSuperfamily: THUMP domain-likeFamily: THUMP domain86 c1jrxA_
not modelled
20.3
46
PDB header: oxidoreductaseChain: A: PDB Molecule: flavocytochrome c;PDBTitle: crystal structure of arg402ala mutant flavocytochrome c32 from shewanella frigidimarina
87 c2v3bB_
not modelled
20.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: rubredoxin 2;PDBTitle: crystal structure of the electron transfer complex2 rubredoxin - rubredoxin reductase from pseudomonas3 aeruginosa.
88 d2rdva_
not modelled
20.0
17
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin89 c2owoA_
not modelled
19.9
17
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
90 c2r7cA_
not modelled
19.7
71
PDB header: rna binding proteinChain: A: PDB Molecule: non-structural rna-binding protein 35;PDBTitle: crystallographic and biochemical analysis of rotavirus nsp22 with nucleotides reveals an ndp kinase like activity
91 d2qifa1
not modelled
19.3
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain92 c2v51E_
not modelled
19.3
58
PDB header: structural protein/contractile proteinChain: E: PDB Molecule: mkl/myocardin-like protein 1;PDBTitle: structure of mal-rpel1 complexed to actin
93 d1hxra_
not modelled
19.1
50
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss494 d2fyma1
not modelled
19.1
23
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase95 c3io1B_
not modelled
19.0
12
PDB header: hydrolaseChain: B: PDB Molecule: aminobenzoyl-glutamate utilization protein;PDBTitle: crystal structure of aminobenzoyl-glutamate utilization2 protein from klebsiella pneumoniae
96 c1ee8A_
not modelled
18.7
24
PDB header: dna binding proteinChain: A: PDB Molecule: mutm (fpg) protein;PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
97 d1tdza3
not modelled
18.6
24
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins98 c2jvaA_
not modelled
18.4
31
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase domain protein;PDBTitle: nmr solution structure of peptidyl-trna hydrolase domain protein from2 pseudomonas syringae pv. tomato. northeast structural genomics3 consortium target psr211
99 d2gmga1
not modelled
18.2
45
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like