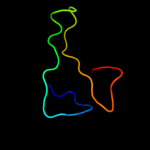
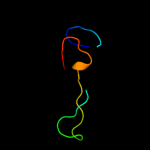
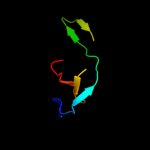
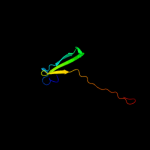
1 c3r1rB_
98.4
19
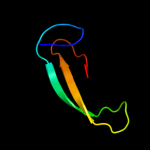
PDB header: complex (oxidoreductase/peptide)Chain: B: PDB Molecule: ribonucleotide reductase r1 protein;PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
2 d1rlra1
97.7
15
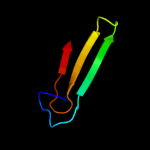
Fold: R1 subunit of ribonucleotide reductase, N-terminal domainSuperfamily: R1 subunit of ribonucleotide reductase, N-terminal domainFamily: R1 subunit of ribonucleotide reductase, N-terminal domain3 d1qypa_
95.4
21
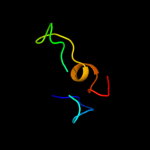
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain4 d1twfi2
94.6
29
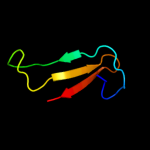
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain5 c3k7aM_
93.8
31
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor iib;PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
6 c1i3qI_
93.3
29
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii 14.2kdPDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
7 d1tfia_
93.0
23
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain8 c3gn5B_
92.9
24
PDB header: dna binding proteinChain: B: PDB Molecule: hth-type transcriptional regulator mqsa (ygit/b3021);PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
9 c2gb5B_
91.6
14
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
10 c3h0gI_
91.3
22
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
11 d1wiia_
89.8
29
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Putative zinc binding domain12 d1dl6a_
86.9
27
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain13 d2fiya1
83.6
20
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like14 d1pfva3
78.2
29
Fold: Rubredoxin-likeSuperfamily: Methionyl-tRNA synthetase (MetRS), Zn-domainFamily: Methionyl-tRNA synthetase (MetRS), Zn-domain15 c2e9hA_
76.6
31
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: solution structure of the eif-5_eif-2b domain from human2 eukaryotic translation initiation factor 5
16 d1rj9b1
75.9
18
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins17 c3hnfA_
75.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large subunit;PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
18 d1ls1a1
74.6
21
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins19 c1nuiA_
73.6
29
PDB header: replicationChain: A: PDB Molecule: dna primase/helicase;PDBTitle: crystal structure of the primase fragment of bacteriophage t7 primase-2 helicase protein
20 c3cngC_
73.3
21
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase;PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
21 c3ndjA_
not modelled
73.2
21
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
22 d1pfta_
not modelled
72.7
31
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain23 d1j8yf1
not modelled
70.6
19
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins24 c2qkdA_
not modelled
70.4
18
PDB header: signaling protein, cell cycleChain: A: PDB Molecule: zinc finger protein zpr1;PDBTitle: crystal structure of tandem zpr1 domains
25 c2elpA_
not modelled
68.9
50
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
26 d1wgwa_
not modelled
68.7
21
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins27 d1vqoz1
not modelled
67.7
46
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae28 c4a17Y_
not modelled
66.5
39
PDB header: ribosomeChain: Y: PDB Molecule: rpl37a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
29 c2zkrz_
not modelled
65.9
33
PDB header: ribosomal protein/rnaChain: Z: PDB Molecule: e site t-rna;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
30 c2qa4Z_
not modelled
65.5
46
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: a more complete structure of the the l7/l12 stalk of the2 haloarcula marismortui 50s large ribosomal subunit
31 c1yshD_
not modelled
65.3
33
PDB header: structural protein/rnaChain: D: PDB Molecule: ribosomal protein l37a;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
32 d2k4xa1
not modelled
64.6
35
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein S27a33 d1jj2y_
not modelled
64.5
25
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae34 c2l7xA_
not modelled
64.5
41
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: crimean congo hemorrhagic fever gn zinc finger
35 c3jyw9_
not modelled
62.8
33
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
36 c1s1i9_
not modelled
62.6
33
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
37 d1ffkw_
not modelled
62.2
25
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae38 c3cc4Z_
not modelled
61.4
48
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: co-crystal structure of anisomycin bound to the 50s ribosomal subunit
39 c2hu9B_
not modelled
54.8
38
PDB header: metal transportChain: B: PDB Molecule: mercuric transport protein periplasmic component;PDBTitle: x-ray structure of the archaeoglobus fulgidus copz n-2 terminal domain
40 d1vd4a_
53.4
42
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain41 d1g2ra_
not modelled
53.2
16
Fold: YlxR-likeSuperfamily: YlxR-likeFamily: YlxR-like42 c2f9iD_
not modelled
53.0
27
PDB header: transferaseChain: D: PDB Molecule: acetyl-coenzyme a carboxylase carboxylPDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
43 d1qzxa1
not modelled
52.3
17
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins44 c3eswA_
not modelled
51.5
23
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
45 c2aklA_
not modelled
50.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
46 c2xigA_
not modelled
50.1
18
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
47 d2cona1
not modelled
50.0
19
Fold: Rubredoxin-likeSuperfamily: NOB1 zinc finger-likeFamily: NOB1 zinc finger-like48 c1x6eA_
not modelled
48.9
15
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger protein 24;PDBTitle: solution structures of the c2h2 type zinc finger domain of2 human zinc finger protein 24
49 c2kvfA_
not modelled
48.5
50
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
50 d1x3za1
not modelled
48.2
10
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core51 c2jq5A_
not modelled
46.6
40
PDB header: structural genomicsChain: A: PDB Molecule: sec-c motif;PDBTitle: solution structure of rpa3114, a sec-c motif containing2 protein from rhodopseudomonas palustris; northeast3 structural genomics consortium target rpt5 / ontario4 center for structural proteomics target rp3097
52 c2kaeA_
not modelled
45.3
18
PDB header: transcription/dnaChain: A: PDB Molecule: gata-type transcription factor;PDBTitle: data-driven model of med1:dna complex
53 c1meyG_
not modelled
45.0
27
PDB header: transferase/dnaChain: G: PDB Molecule: consensus zinc finger;PDBTitle: crystal structure of a designed zinc finger protein bound2 to dna
54 c2dmiA_
not modelled
43.9
7
PDB header: transcriptionChain: A: PDB Molecule: teashirt homolog 3;PDBTitle: solution structure of the first and the second zf-c2h2 like2 domains of human teashirt homolog 3
55 c2jneA_
not modelled
43.4
40
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
56 d2jnea1
not modelled
43.4
40
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like57 c2f9yB_
not modelled
43.3
21
PDB header: ligaseChain: B: PDB Molecule: acetyl-coenzyme a carboxylase carboxyl transferase subunitPDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
58 d2f9yb1
not modelled
43.3
21
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain59 c2ctdA_
not modelled
42.9
23
PDB header: metal binding proteinChain: A: PDB Molecule: zinc finger protein 512;PDBTitle: solution structure of two zf-c2h2 domains from human zinc2 finger protein 512
60 c2cotA_
not modelled
42.4
15
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 435;PDBTitle: solution structure of the first and second zf-c2h2 domain2 of zinc finger protein 435
61 d1tlqa_
not modelled
42.4
20
Fold: YutG-likeSuperfamily: YutG-likeFamily: YutG-like62 c1tlqA_
not modelled
42.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ypjq;PDBTitle: crystal structure of protein ypjq from bacillus subtilis, pfam duf64
63 d2cota1
not modelled
42.2
30
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H264 d1x6ea2
not modelled
42.1
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H265 c3hi2C_
not modelled
41.6
57
PDB header: dna binding protein/toxinChain: C: PDB Molecule: hth-type transcriptional regulator mqsa(ygit);PDBTitle: structure of the n-terminal domain of the e. coli antitoxin mqsa2 (ygit/b3021) in complex with the e. coli toxin mqsr (ygiu/b3022)
66 c3c8fA_
not modelled
41.2
45
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
67 c3g9yA_
not modelled
41.0
30
PDB header: transcription/rnaChain: A: PDB Molecule: zinc finger ran-binding domain-containing protein 2;PDBTitle: crystal structure of the second zinc finger from zranb2/znf265 bound2 to 6 nt ssrna sequence agguaa
68 d1a1ia2
not modelled
40.8
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H269 c1jocA_
not modelled
40.8
36
PDB header: membrane proteinChain: A: PDB Molecule: early endosomal autoantigen 1;PDBTitle: eea1 homodimer of c-terminal fyve domain bound to inositol2 1,3-diphosphate
70 d1llmc2
not modelled
40.8
30
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H271 d1srka_
not modelled
39.5
45
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H272 c3zyqA_
not modelled
39.2
25
PDB header: signalingChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
73 d1u85a1
not modelled
38.6
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H274 c2gqjA_
not modelled
38.1
20
PDB header: gene regulationChain: A: PDB Molecule: zinc finger protein kiaa1196;PDBTitle: solution structure of the two zf-c2h2 like domains(493-575)2 of human zinc finger protein kiaa1196
75 c1rikA_
not modelled
38.0
40
PDB header: de novo proteinChain: A: PDB Molecule: e6apc1 peptide;PDBTitle: e6-binding zinc finger (e6apc1)
76 d1p91a_
not modelled
37.6
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase RlmA77 d1xf7a_
not modelled
37.4
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H278 c1xf7A_
not modelled
37.4
40
PDB header: transcriptionChain: A: PDB Molecule: wilms' tumor protein;PDBTitle: high resolution nmr structure of the wilms' tumor2 suppressor protein (wt1) finger 3
79 c2k2dA_
not modelled
36.7
33
PDB header: metal binding proteinChain: A: PDB Molecule: ring finger and chy zinc finger domain-PDBTitle: solution nmr structure of c-terminal domain of human pirh2.2 northeast structural genomics consortium (nesg) target ht2c
80 d1m2ka_
not modelled
36.3
14
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators81 d1x6ea1
not modelled
36.3
27
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H282 c2elsA_
not modelled
36.3
36
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 2nd c2h2 zinc finger of human2 zinc finger protein 406
83 c2lk0A_
not modelled
36.0
37
PDB header: rna binding proteinChain: A: PDB Molecule: rna-binding protein 5;PDBTitle: solution structure and binding studies of the ranbp2-type zinc finger2 of rbm5
84 c1areA_
not modelled
35.2
40
PDB header: transcription regulationChain: A: PDB Molecule: yeast transcription factor adr1;PDBTitle: structures of dna-binding mutant zinc finger domains:2 implications for dna binding
85 c3lpeF_
not modelled
34.5
30
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
86 d1p7aa_
not modelled
34.3
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H287 c2epsA_
not modelled
33.9
18
PDB header: transcriptionChain: A: PDB Molecule: poz-, at hook-, and zinc finger-containingPDBTitle: solution structure of the 4th zinc finger domain of zinc2 finger protein 278
88 d1a1ia3
not modelled
33.6
30
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H289 d1joca1
not modelled
33.6
35
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain90 c2jrpA_
not modelled
33.6
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
91 c1dgsB_
not modelled
33.0
48
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
92 c1ltlE_
not modelled
32.9
16
PDB header: replicationChain: E: PDB Molecule: dna replication initiator (cdc21/cdc54);PDBTitle: the dodecamer structure of mcm from archaeal m.2 thermoautotrophicum
93 c2ztgA_
not modelled
32.9
22
PDB header: ligaseChain: A: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of archaeoglobus fulgidus alanyl-trna2 synthetase lacking the c-terminal dimerization domain in3 complex with ala-sa
94 d1lkoa2
not modelled
32.8
24
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin95 c1va3A_
not modelled
32.8
40
PDB header: transcriptionChain: A: PDB Molecule: transcription factor sp1;PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 3)
96 d2dula1
not modelled
32.1
11
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TRM1-like97 c1sp1A_
not modelled
32.0
40
PDB header: zinc fingerChain: A: PDB Molecule: sp1f3;PDBTitle: nmr structure of a zinc finger domain from transcription2 factor sp1f3, minimized average structure
98 d1sp1a_
not modelled
32.0
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H299 c2hl7A_
not modelled
31.9
40
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
100 d2j0151
not modelled
31.9
29
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p101 c1v9pB_
not modelled
31.8
48
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
102 c2l8eA_
not modelled
31.8
28
PDB header: dna binding proteinChain: A: PDB Molecule: polyhomeotic-like protein 1;PDBTitle: solution nmr structure of fcs domain of human polyhomeotic homolog 12 (hph1)
103 c3f2cA_
not modelled
31.6
25
PDB header: transferase/dnaChain: A: PDB Molecule: geobacillus kaustophilus dna polc;PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
104 c3floD_
not modelled
31.6
24
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
105 d2zjrz1
not modelled
31.5
24
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p106 c1z2qA_
not modelled
31.4
31
PDB header: membrane proteinChain: A: PDB Molecule: lm5-1;PDBTitle: high-resolution solution structure of the lm5-1 fyve domain2 from leishmania major
107 c2kw0A_
not modelled
31.4
30
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
108 c3pkiF_
not modelled
31.3
21
PDB header: hydrolaseChain: F: PDB Molecule: nad-dependent deacetylase sirtuin-6;PDBTitle: human sirt6 crystal structure in complex with adp ribose
109 c1rimA_
not modelled
31.2
40
PDB header: de novo proteinChain: A: PDB Molecule: e6apc2 peptide;PDBTitle: e6-binding zinc finger (e6apc2)
110 c2yteA_
not modelled
31.1
36
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 473;PDBTitle: solution structure of the c2h2 type zinc finger (region 484-2 512) of human zinc finger protein 473
111 d2gmga1
not modelled
31.0
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like112 d1enwa_
not modelled
31.0
10
Fold: RuvA C-terminal domain-likeSuperfamily: Elongation factor TFIIS domain 2Family: Elongation factor TFIIS domain 2113 c1ardA_
not modelled
30.8
40
PDB header: transcription regulationChain: A: PDB Molecule: yeast transcription factor adr1;PDBTitle: structures of dna-binding mutant zinc finger domains:2 implications for dna binding
114 d2adra1
not modelled
30.6
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2115 d2epqa1
not modelled
30.4
33
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2116 d1ltla_
not modelled
30.2
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA replication initiator (cdc21/cdc54) N-terminal domain117 c2eltA_
not modelled
30.1
18
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 3rd c2h2 zinc finger of human2 zinc finger protein 406
118 d1k81a_
not modelled
29.8
50
Fold: Zinc-binding domain of translation initiation factor 2 betaSuperfamily: Zinc-binding domain of translation initiation factor 2 betaFamily: Zinc-binding domain of translation initiation factor 2 beta119 d2ct1a2
not modelled
29.8
45
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2120 d1ubdc2
not modelled
29.2
31
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2