1 c3qnqD_
100.0
15
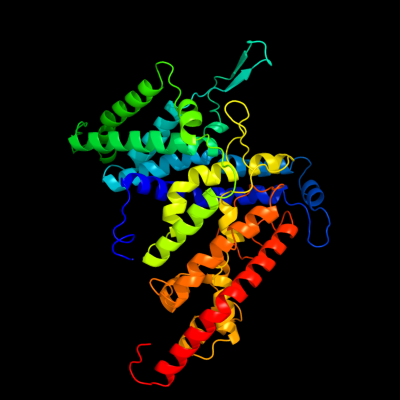
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
2 d3bp8c1
99.9
41
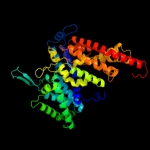
Fold: Homing endonuclease-likeSuperfamily: Glucose permease domain IIBFamily: Glucose permease domain IIB3 c1ibaA_
99.9
41
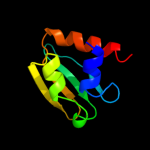
PDB header: phoshphotransferaseChain: A: PDB Molecule: glucose permease;PDBTitle: glucose permease (domain iib), nmr, 11 structures
4 c3ipjB_
99.8
28
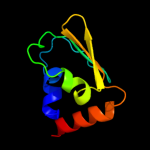
PDB header: transferaseChain: B: PDB Molecule: pts system, iiabc component;PDBTitle: the crystal structure of one domain of the pts system, iiabc component2 from clostridium difficile
5 c3mp7A_
47.0
16
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase subunit secy;PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
6 d1xmea1
45.8
13
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like7 c3b9yA_
39.8
13
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
8 c2hnhA_
38.1
35
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii alpha subunit;PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
9 d1gpra_
37.8
8
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like10 c2dlaB_
36.9
29
PDB header: replicationChain: B: PDB Molecule: 397aa long hypothetical protein;PDBTitle: primase large subunit amino terminal domain from pyrococcus horikoshii
11 d2gpra_
33.6
3
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like12 d1glaf_
27.6
5
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like13 d2f3ga_
27.6
9
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like14 c3e0dA_
25.9
35
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase iii subunit alpha;PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
15 c2rddB_
25.7
39
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
16 d1nh8a2
24.9
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain17 c3zqsB_
18.8
35
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase fancl;PDBTitle: human fancl central domain
18 d2nr9a1
18.5
17
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like19 d1h3da2
16.6
24
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain20 d1l0wa3
16.2
14
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain21 d1osda_
not modelled
15.2
17
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain22 d1cpza_
not modelled
14.9
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain23 d1st6a5
not modelled
14.6
19
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin24 c2ci2I_
not modelled
14.1
30
PDB header: proteinase inhibitor (chymotrypsin)Chain: I: PDB Molecule: chymotrypsin inhibitor 2;PDBTitle: crystal and molecular structure of the serine proteinase2 inhibitor ci-2 from barley seeds
25 c1p68A_
not modelled
14.1
55
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed protein s-824;PDBTitle: solution structure of s-824, a de novo designed four helix2 bundle
26 d2cfua2
not modelled
13.2
19
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Alkylsulfatase-like27 d1q8la_
not modelled
13.0
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain28 d1kvja_
not modelled
12.6
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain29 c2ga7A_
not modelled
12.6
21
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
30 d1fzda_
not modelled
12.4
29
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like31 d1j7na3
not modelled
12.1
15
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins32 c2ldiA_
not modelled
11.7
19
PDB header: hydrolaseChain: A: PDB Molecule: zinc-transporting atpase;PDBTitle: nmr solution structure of ziaan sub mutant
33 d2axtk1
not modelled
11.4
40
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein K, PsbKFamily: PsbK-like34 c2cfuA_
not modelled
11.2
19
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
35 c1yjrA_
not modelled
11.1
12
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
36 c3f2cA_
not modelled
10.6
26
PDB header: transferase/dnaChain: A: PDB Molecule: geobacillus kaustophilus dna polc;PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
37 d1u7ga_
not modelled
10.3
3
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter38 d2ifqa1
not modelled
10.3
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase39 d1f9qa_
not modelled
10.1
10
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines40 c2w2eA_
not modelled
10.1
7
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin,2 aqy1, in a closed conformation at ph 3.5
41 d2a9da1
not modelled
10.0
43
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Molybdenum-containing oxidoreductases-like dimerisation domain42 d1p6ta1
not modelled
9.7
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain43 c3a0bK_
not modelled
9.7
33
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of br-substituted photosystem ii complex
44 c3a0bk_
not modelled
9.7
33
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of br-substituted photosystem ii complex
45 d2qifa1
not modelled
9.2
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain46 c2l3mA_
not modelled
9.0
17
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
47 d3pvia_
not modelled
9.0
16
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease PvuII48 d1afia_
not modelled
9.0
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain49 c3e6qL_
not modelled
8.9
24
PDB header: isomeraseChain: L: PDB Molecule: putative 5-carboxymethyl-2-hydroxymuconate isomerase;PDBTitle: putative 5-carboxymethyl-2-hydroxymuconate isomerase from pseudomonas2 aeruginosa.
50 c3a0hk_
not modelled
8.7
33
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of i-substituted photosystem ii complex
51 d1pfma_
not modelled
8.5
10
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines52 d1fida_
not modelled
8.3
24
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like53 d2axti1
not modelled
7.8
23
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like54 d1plfa_
not modelled
7.7
10
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines55 c3eh4A_
not modelled
7.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase subunit 1;PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
56 c1vbwA_
not modelled
7.5
24
PDB header: protein bindingChain: A: PDB Molecule: trypsin inhibitor bgit;PDBTitle: crystal structure of bitter gourd trypsin inhibitor
57 d1lwuc1
not modelled
7.5
31
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like58 d1wf3a2
not modelled
7.4
21
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)59 d1ypci_
not modelled
7.4
29
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors60 d1f9ra_
not modelled
7.2
10
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines61 c3l7qD_
not modelled
7.2
16
PDB header: translationChain: D: PDB Molecule: putative translation initiation inhibitor, aldr regulator-PDBTitle: crystal structure of aldr from streptococcus mutans
62 d1to2i_
not modelled
7.0
29
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors63 c1q1kA_
not modelled
7.0
17
PDB header: transferaseChain: A: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: structure of atp-phosphoribosyltransferase from e. coli complexed with2 pr-atp
64 d1h7sa2
not modelled
6.9
37
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain65 c1u5tB_
not modelled
6.9
14
PDB header: transport proteinChain: B: PDB Molecule: defective in vacuolar protein sorting; vps36p;PDBTitle: structure of the escrt-ii endosomal trafficking complex
66 c1hgvA_
not modelled
6.9
10
PDB header: virusChain: A: PDB Molecule: ph75 inovirus major coat protein;PDBTitle: filamentous bacteriophage ph75
67 d2snii_
not modelled
6.9
30
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors68 c3dxsX_
not modelled
6.8
19
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
69 d1z9ha1
not modelled
6.8
17
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain70 d1dwma_
not modelled
6.7
31
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors71 c1y3kA_
not modelled
6.6
14
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
72 c1tinA_
not modelled
6.6
35
PDB header: serine protease inhibitorChain: A: PDB Molecule: trypsin inhibitor v;PDBTitle: three-dimensional structure in solution of cucurbita maxima2 trypsin inhibitor-v determined by nmr spectroscopy
73 d1bkna2
not modelled
6.6
26
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain74 d1s6ua_
not modelled
6.5
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain75 d1l0wa2
not modelled
6.4
16
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain76 c3hd7A_
not modelled
6.4
15
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
77 d1egla_
not modelled
6.3
18
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors78 c3qd1X_
not modelled
6.3
26
PDB header: sugar binding proteinChain: X: PDB Molecule: platelet binding protein gspb;PDBTitle: gspb plus alpha-2,3-sialyl (1-thioethyl)galactose
79 c2j61B_
not modelled
6.2
35
PDB header: lectinChain: B: PDB Molecule: ficolin-2;PDBTitle: l-ficolin complexed to n-acetylglucosamine (forme c)
80 d1lwub1
not modelled
6.2
19
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like81 d1m1jc1
not modelled
6.2
30
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like82 d1wffa_
not modelled
6.1
30
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger83 d1xrga_
not modelled
6.1
16
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP84 d2aw0a_
not modelled
6.1
17
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain85 d1wgla_
not modelled
6.0
21
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: CUE domain86 c2hqnA_
not modelled
6.0
21
PDB header: signaling proteinChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
87 c1yg0A_
not modelled
5.9
14
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
88 c3cjsA_
not modelled
5.9
15
PDB header: transferase/ribosomal proteinChain: A: PDB Molecule: ribosomal protein l11 methyltransferase;PDBTitle: minimal recognition complex between prma and ribosomal protein l11
89 c2ofhX_
not modelled
5.8
17
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
90 c1a0oH_
not modelled
5.7
15
PDB header: chemotaxisChain: H: PDB Molecule: chea;PDBTitle: chey-binding domain of chea in complex with chey
91 d1jmkc_
not modelled
5.7
9
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases92 c2kncA_
not modelled
5.7
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
93 c1deqF_
not modelled
5.6
24
PDB header: PDB COMPND: 94 c1w7pD_
not modelled
5.6
14
PDB header: protein transportChain: D: PDB Molecule: vps36p, ylr417w;PDBTitle: the crystal structure of endosomal complex escrt-ii2 (vps22/vps25/vps36)
95 d1qaha_
not modelled
5.6
23
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP96 d1p6ta2
not modelled
5.6
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain97 c1b9uA_
not modelled
5.6
22
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
98 c3bvhC_
not modelled
5.5
24
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
99 c2hpcF_
not modelled
5.5
24
PDB header: blood clottingChain: F: PDB Molecule: fibrinogen, gamma polypeptide;PDBTitle: crystal structure of fragment d from human fibrinogen complexed with2 gly-pro-arg-pro-amide.