1 c3ne8A_
100.0
38
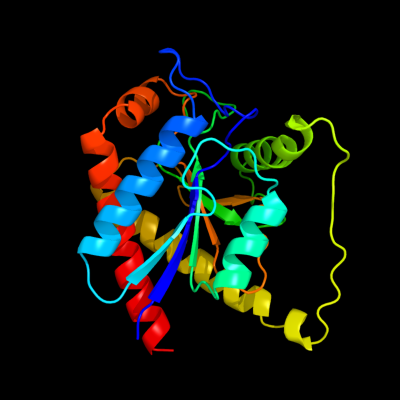
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of a domain from n-acetylmuramoyl-l-alanine2 amidase of bartonella henselae str. houston-1
2 d1jwqa_
100.0
35
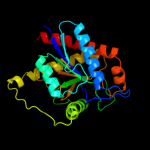
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: N-acetylmuramoyl-L-alanine amidase-like3 c1xovA_
100.0
16
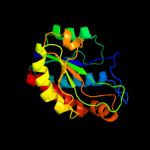
PDB header: hydrolaseChain: A: PDB Molecule: ply protein;PDBTitle: the crystal structure of the listeria monocytogenes bacteriophage psa2 endolysin plypsa
4 c3czxA_
100.0
21
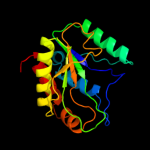
PDB header: hydrolaseChain: A: PDB Molecule: putative n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of the putative n-acetylmuramoyl-l-2 alanine amidase from neisseria meningitidis
5 d1xova2
100.0
19
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: N-acetylmuramoyl-L-alanine amidase-like6 c3qayC_
100.0
25
PDB header: lyaseChain: C: PDB Molecule: endolysin;PDBTitle: catalytic domain of cd27l endolysin targeting clostridia difficile
7 d2gfqa1
87.2
21
Fold: Phosphorylase/hydrolase-likeSuperfamily: AF0625-likeFamily: AF0625-like8 c2gfqC_
85.0
25
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: upf0204 protein ph0006;PDBTitle: structure of protein of unknown function ph0006 from pyrococcus2 horikoshii
9 c2qvpC_
84.2
15
PDB header: hydrolaseChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative metallopeptidase (sama_0725) from2 shewanella amazonensis sb2b at 2.00 a resolution
10 d1yqea1
82.4
33
Fold: Phosphorylase/hydrolase-likeSuperfamily: AF0625-likeFamily: AF0625-like11 c1c4gB_
61.5
18
PDB header: transferaseChain: B: PDB Molecule: protein (alpha-d-glucose 1-phosphatePDBTitle: phosphoglucomutase vanadate based transition state analog2 complex
12 c2fuvB_
58.8
19
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucomutase;PDBTitle: phosphoglucomutase from salmonella typhimurium.
13 c1wqaB_
47.4
29
PDB header: isomeraseChain: B: PDB Molecule: phospho-sugar mutase;PDBTitle: crystal structure of pyrococcus horikoshii2 phosphomannomutase/phosphoglucomutase complexed with mg2+
14 d1nyra1
46.0
18
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS15 c2f7lA_
44.3
24
PDB header: isomeraseChain: A: PDB Molecule: 455aa long hypothetical phospho-sugar mutase;PDBTitle: crystal structure of sulfolobus tokodaii2 phosphomannomutase/phosphoglucomutase
16 c1wwpA_
43.3
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha0636;PDBTitle: crystal structure of ttk003001694 from thermus thermophilus2 hb8
17 d1fx0b1
40.8
15
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase18 c3i3wB_
40.6
22
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucosamine mutase;PDBTitle: structure of a phosphoglucosamine mutase from francisella tularensis
19 d1kfia2
38.5
21
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains20 c3l2nA_
35.9
18
PDB header: hydrolaseChain: A: PDB Molecule: peptidase m14, carboxypeptidase a;PDBTitle: crystal structure of putative carboxypeptidase a (yp_562911.1) from2 shewanella denitrificans os-217 at 2.39 a resolution
21 d1skye1
not modelled
35.8
18
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase22 d3pmga2
not modelled
34.2
14
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains23 c3nh8A_
not modelled
33.7
15
PDB header: hydrolaseChain: A: PDB Molecule: aspartoacylase-2;PDBTitle: crystal structure of murine aminoacylase 3 in complex with n-acetyl-s-2 1,2-dichlorovinyl-l-cysteine
24 c3r1jB_
not modelled
31.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: alpha-ketoglutarate-dependent taurine dioxygenase;PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
25 d1ad1a_
not modelled
29.9
75
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase26 c1nj2A_
not modelled
29.5
23
PDB header: ligaseChain: A: PDB Molecule: proline-trna synthetase;PDBTitle: crystal structure of prolyl-trna synthetase from methanothermobacter2 thermautotrophicus
27 d2g4ca1
not modelled
29.4
15
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS28 c3k2kA_
not modelled
29.2
18
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxypeptidase;PDBTitle: crystal structure of putative carboxypeptidase (yp_103406.1) from2 burkholderia mallei atcc 23344 at 2.49 a resolution
29 c3bghB_
not modelled
27.6
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative neuraminyllactose-binding hemagglutinin homolog;PDBTitle: crystal structure of putative neuraminyllactose-binding hemagglutinin2 homolog from helicobacter pylori
30 d1wu7a1
not modelled
24.5
18
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS31 d1nj8a1
not modelled
24.3
16
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS32 d1nj1a1
not modelled
23.8
22
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS33 c2y5sA_
not modelled
23.8
63
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
34 d1qf6a1
not modelled
23.0
7
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS35 d1g5ha1
not modelled
22.4
17
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS36 c3l80A_
not modelled
21.8
14
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein smu.1393c;PDBTitle: crystal structure of smu.1393c from streptococcus mutans ua159
37 c2dzaA_
not modelled
21.5
63
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
38 d1p5dx2
not modelled
21.5
34
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains39 d1kija1
not modelled
20.2
13
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain40 d1u83a_
not modelled
18.9
24
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA41 c1u83A_
not modelled
18.9
24
PDB header: lyaseChain: A: PDB Molecule: phosphosulfolactate synthase;PDBTitle: psl synthase from bacillus subtilis
42 d1eyea_
not modelled
18.7
44
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase43 c1kfiA_
not modelled
18.4
21
PDB header: isomeraseChain: A: PDB Molecule: phosphoglucomutase 1;PDBTitle: crystal structure of the exocytosis-sensitive2 phosphoprotein, pp63/parafusin (phosphoglucomutase) from3 paramecium
44 c2vp8A_
not modelled
18.1
38
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase 2;PDBTitle: structure of mycobacterium tuberculosis rv1207
45 c2ae3A_
not modelled
17.9
14
PDB header: hydrolaseChain: A: PDB Molecule: glutaryl 7-aminocephalosporanic acid acylase;PDBTitle: glutaryl 7-aminocephalosporanic acid acylase: mutational study of2 activation mechanism
46 c2ronA_
not modelled
17.8
15
PDB header: hydrolaseChain: A: PDB Molecule: surfactin synthetase thioesterase subunit;PDBTitle: the external thioesterase of the surfactin-synthetase
47 c2gx8B_
not modelled
17.8
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
48 d1tx2a_
not modelled
17.8
47
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase49 c1tx2A_
not modelled
17.8
47
PDB header: transferaseChain: A: PDB Molecule: dhps, dihydropteroate synthase;PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
50 d2b3ya2
not modelled
17.5
20
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain51 c2e85B_
not modelled
17.3
17
PDB header: hydrolaseChain: B: PDB Molecule: hydrogenase 3 maturation protease;PDBTitle: crystal structure of the hydrogenase 3 maturation protease
52 c3ibtA_
not modelled
16.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase;PDBTitle: structure of 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (qdo)
53 d1ei1a1
not modelled
15.7
19
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain54 c1k8wA_
not modelled
15.6
55
PDB header: lyase/rnaChain: A: PDB Molecule: trna pseudouridine synthase b;PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
55 d1ccwa_
not modelled
15.4
17
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain56 c2vf7B_
not modelled
15.4
28
PDB header: dna-binding proteinChain: B: PDB Molecule: excinuclease abc, subunit a.;PDBTitle: crystal structure of uvra2 from deinococcus radiodurans
57 d2gx8a1
not modelled
14.7
23
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like58 c1ze2B_
not modelled
14.5
58
PDB header: lyase/rnaChain: B: PDB Molecule: trna pseudouridine synthase b;PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
59 c3netB_
not modelled
14.2
15
PDB header: ligaseChain: B: PDB Molecule: histidyl-trna synthetase;PDBTitle: crystal structure of histidyl-trna synthetase from nostoc sp. pcc 7120
60 c3t4cD_
not modelled
14.0
21
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 1;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
61 d1qe0a1
not modelled
13.7
13
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS62 c3cwvB_
not modelled
13.7
3
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase, b subunit, truncated;PDBTitle: crystal structure of b-subunit of the dna gyrase from myxococcus2 xanthus
63 c2qj8B_
not modelled
13.6
19
PDB header: hydrolaseChain: B: PDB Molecule: mlr6093 protein;PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
64 d3bula2
not modelled
13.4
17
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain65 c2i4lC_
not modelled
13.2
11
PDB header: ligaseChain: C: PDB Molecule: proline-trna ligase;PDBTitle: rhodopseudomonas palustris prolyl-trna synthetase
66 c5acnA_
not modelled
13.2
20
PDB header: lyase(carbon-oxygen)Chain: A: PDB Molecule: aconitase;PDBTitle: structure of activated aconitase. formation of the (4fe-4s)2 cluster in the crystal
67 d1c4xa_
not modelled
13.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase68 c2yxbA_
not modelled
12.9
18
PDB header: isomeraseChain: A: PDB Molecule: coenzyme b12-dependent mutase;PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
69 d1acoa2
not modelled
12.8
20
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain70 d2bodx1
not modelled
12.7
29
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases71 d2fsja1
not modelled
12.4
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Ta0583-like72 d1joga_
not modelled
12.3
25
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit73 d1cr5a2
not modelled
12.2
21
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like74 c2wj4B_
not modelled
11.9
26
PDB header: oxidoreductaseChain: B: PDB Molecule: 1h-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase;PDBTitle: crystal structure of the cofactor-devoid 1-h-3-hydroxy-4-2 oxoquinaldine 2,4-dioxygenase (hod) from arthrobacter3 nitroguajacolicus ru61a anaerobically complexed with its4 natural substrate 1-h-3-hydroxy-4-oxoquinaldine
75 d2ey4a2
not modelled
11.8
55
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB76 c1g5hA_
not modelled
11.5
17
PDB header: dna binding proteinChain: A: PDB Molecule: mitochondrial dna polymerase accessory subunit;PDBTitle: crystal structure of the accessory subunit of murine mitochondrial2 polymerase gamma
77 d1kjna_
not modelled
11.4
20
Fold: Hypothetical protein MTH777 (MT0777)Superfamily: Hypothetical protein MTH777 (MT0777)Family: Hypothetical protein MTH777 (MT0777)78 d1ifya_
not modelled
11.3
12
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain79 c3cdxB_
not modelled
11.2
20
PDB header: hydrolaseChain: B: PDB Molecule: succinylglutamatedesuccinylase/aspartoacylase;PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
80 c3lnuA_
not modelled
11.1
6
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase iv subunit b;PDBTitle: crystal structure of pare subunit
81 d2fywa1
not modelled
10.9
28
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like82 c3dyvA_
not modelled
10.9
31
PDB header: hydrolaseChain: A: PDB Molecule: esterase d;PDBTitle: snapshots of esterase d from lactobacillus rhamnosus:2 insights into a rotation driven catalytic mechanism
83 c2b3yB_
not modelled
10.9
20
PDB header: lyaseChain: B: PDB Molecule: iron-responsive element binding protein 1;PDBTitle: structure of a monoclinic crystal form of human cytosolic aconitase2 (irp1)
84 d1sgva2
not modelled
10.8
55
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB85 d1uoza_
not modelled
10.4
31
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases86 d1uuqa_
not modelled
10.3
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases87 c1uz4A_
not modelled
10.3
14
PDB header: hydrolaseChain: A: PDB Molecule: man5a;PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
88 d2apoa2
not modelled
10.3
55
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB89 d1k8wa5
not modelled
10.0
55
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB90 d1r3ea2
not modelled
10.0
64
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase II TruB91 c3qvmA_
not modelled
10.0
20
PDB header: hydrolaseChain: A: PDB Molecule: olei00960;PDBTitle: the structure of olei00960, a hydrolase from oleispira antarctica
92 c2dlnA_
not modelled
9.9
18
PDB header: ligase(peptidoglycan synthesis)Chain: A: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: vancomycin resistance: structure of d-alanine:d-alanine2 ligase at 2.3 angstroms resolution
93 c3a64A_
not modelled
9.8
38
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: crystal structure of cccel6c, a glycoside hydrolase family 62 enzyme, from coprinopsis cinerea
94 c2i9oA_
not modelled
9.7
21
PDB header: de novo proteinChain: A: PDB Molecule: mhb8a peptide;PDBTitle: design of bivalent miniprotein consisting of two2 independent elements, a b-hairpin peptide and a-helix3 peptide, tethered by eight glycines
95 c2daiA_
not modelled
9.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubiquitin associated domain containing 1;PDBTitle: solution structure of the first uba domain in the human2 ubiquitin associated domain containing 1 (ubadc1)
96 c1y37A_
not modelled
9.5
20
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: structure of fluoroacetate dehalogenase from burkholderia sp. fa1
97 c1y80A_
not modelled
9.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted cobalamin binding protein;PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
98 d1s16a1
not modelled
9.4
10
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain99 c3pdkB_
not modelled
9.3
17
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucosamine mutase;PDBTitle: crystal structure of phosphoglucosamine mutase from b. anthracis