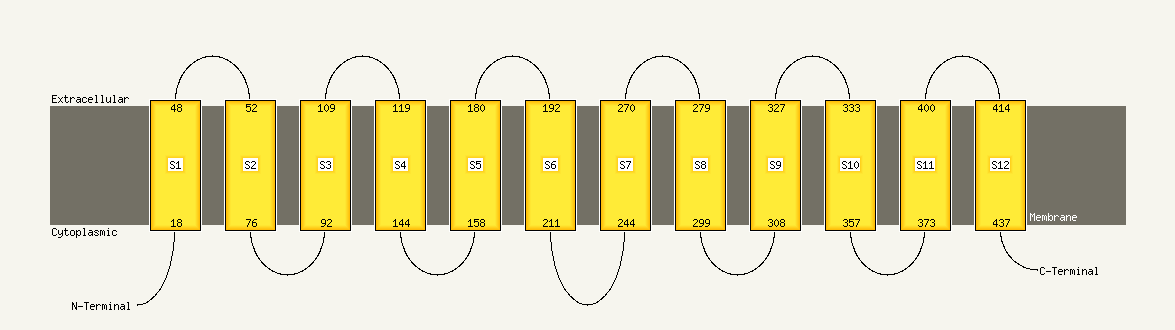
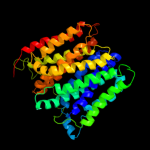
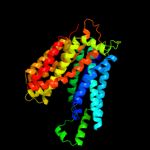
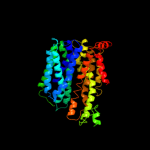
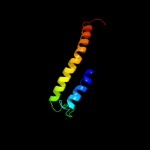
1 d1pw4a_
100.0
11
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter2 d1pv7a_
100.0
14
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter3 c2gfpA_
99.9
11
PDB header: membrane proteinChain: A: PDB Molecule: multidrug resistance protein d;PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
4 c3o7pA_
99.9
12
PDB header: transport proteinChain: A: PDB Molecule: l-fucose-proton symporter;PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
5 c2xutC_
99.9
11
PDB header: transport proteinChain: C: PDB Molecule: proton/peptide symporter family protein;PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
6 c3qnqD_
60.1
14
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
7 c1m46B_
17.4
25
PDB header: cell cycle proteinChain: B: PDB Molecule: iq4 motif from myo2p, a class v myosin;PDBTitle: crystal structure of mlc1p bound to iq4 of myo2p, a class v2 myosin
8 c2kncB_
15.7
14
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
9 c2rdcA_
13.6
15
PDB header: lipid binding proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
10 c1l4aE_
13.4
24
PDB header: endocytosis/exocytosisChain: E: PDB Molecule: synaphin a;PDBTitle: x-ray structure of the neuronal complexin/snare complex2 from the squid loligo pealei
11 c1by0A_
10.8
13
PDB header: rna binding proteinChain: A: PDB Molecule: protein (hepatitis delta antigen);PDBTitle: n-terminal leucine-repeat region of hepatitis delta antigen
12 c3d9sB_
9.8
16
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin-5;PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
13 d1sb0a_
9.0
27
Fold: Kix domain of CBP (creb binding protein)Superfamily: Kix domain of CBP (creb binding protein)Family: Kix domain of CBP (creb binding protein)14 d2p0ma1
8.9
24
Fold: LipoxigenaseSuperfamily: LipoxigenaseFamily: Animal lipoxigenases15 d2b5ua2
7.9
14
Fold: Cloacin translocation domainSuperfamily: Cloacin translocation domainFamily: Cloacin translocation domain16 c2rddB_
7.6
10
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
17 c3d3lB_
6.8
24
PDB header: oxidoreductaseChain: B: PDB Molecule: arachidonate 12-lipoxygenase, 12s-type;PDBTitle: the 2.6 a crystal structure of the lipoxygenase domain of2 human arachidonate 12-lipoxygenase, 12s-type (casp target)
18 c2fnqB_
6.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: allene oxide synthase-lipoxygenase protein;PDBTitle: insights from the x-ray crystal structure of coral 8r-2 lipoxygenase: calcium activation via a c2-like domain and3 a structural basis of product chirality
19 c1c94B_
6.3
12
PDB header: gene regulationChain: B: PDB Molecule: retro-gcn4 leucine zipper;PDBTitle: reversing the sequence of the gcn4 leucine zipper does not2 affect its fold.
20 c2kjeB_
6.2
36
PDB header: transcriptionChain: B: PDB Molecule: early e1a 32 kda protein;PDBTitle: nmr structure of cbp taz2 and adenoviral e1a complex
21 c2p0mB_
not modelled
6.2
24
PDB header: oxidoreductaseChain: B: PDB Molecule: arachidonate 15-lipoxygenase;PDBTitle: revised structure of rabbit reticulocyte 15s-lipoxygenase
22 c2oarA_
not modelled
6.2
6
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
23 d1ymga1
not modelled
6.2
8
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like24 c1ymgA_
not modelled
6.2
8
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
25 c3o8yA_
not modelled
6.1
29
PDB header: oxidoreductaseChain: A: PDB Molecule: arachidonate 5-lipoxygenase;PDBTitle: stable-5-lipoxygenase
26 c3mkuA_
not modelled
5.7
10
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
27 d1j4na_
not modelled
5.7
9
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like28 c1a92B_
not modelled
5.6
13
PDB header: leucine zipperChain: B: PDB Molecule: delta antigen;PDBTitle: oligomerization domain of hepatitis delta antigen
29 c2w8aC_
not modelled
5.4
8
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
30 c2hjmB_
not modelled
5.4
8
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein pf1176;PDBTitle: crystal structure of a singleton protein pf1176 from p. furiosus
31 c2kmfA_
not modelled
5.3
25
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kda protein;PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
32 c3r4hD_
not modelled
5.3
28
PDB header: de novo proteinChain: D: PDB Molecule: coiled coil helix cc-tet-phi22;PDBTitle: crystal structure of the 4-helix coiled coil cc-tet-phi22
33 c3r4hE_
not modelled
5.0
28
PDB header: de novo proteinChain: E: PDB Molecule: coiled coil helix cc-tet-phi22;PDBTitle: crystal structure of the 4-helix coiled coil cc-tet-phi22