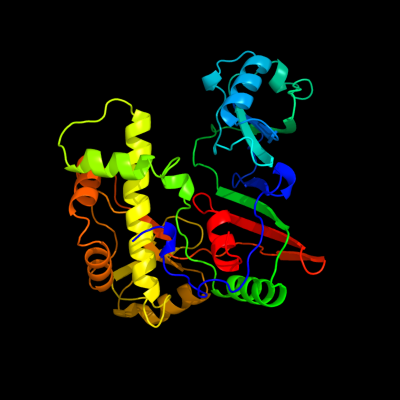
| 1 | c1sb7A_
|
|
 |
100.0 |
99 |
PDB header:lyase
Chain: A: PDB Molecule:trna pseudouridine synthase d;
PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
|
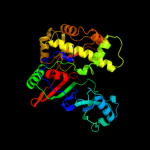
| 2 | d1szwa_
|
|
 |
100.0 |
99 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:tRNA pseudouridine synthase TruD |
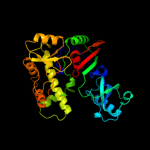
| 3 | c1z2zB_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:probable trna pseudouridine synthase d;
PDBTitle: crystal structure of the putative trna pseudouridine2 synthase d (trud) from methanosarcina mazei, northeast3 structural genomics target mar1
|
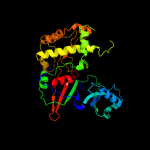
| 4 | d1k8wa5
|
|
 |
86.5 |
12 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 5 | d1r3ea2
|
|
 |
84.1 |
16 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 6 | d2cx6a1
|
|
 |
83.6 |
16 |
Fold:Barstar-like
Superfamily:Barstar-related
Family:Barstar-related |
| 7 | c1sgvA_
|
|
 |
81.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: structure of trna psi55 pseudouridine synthase (trub)
|
| 8 | d2ey4a2
|
|
 |
81.6 |
17 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 9 | d2apoa2
|
|
 |
78.3 |
16 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 10 | d1sgva2
|
|
 |
75.0 |
15 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 11 | c1k8wA_
|
|
 |
72.8 |
11 |
PDB header:lyase/rna
Chain: A: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
|
| 12 | c1ze2B_
|
|
 |
68.4 |
33 |
PDB header:lyase/rna
Chain: B: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
|
| 13 | d1ay7b_
|
|
 |
62.7 |
20 |
Fold:Barstar-like
Superfamily:Barstar-related
Family:Barstar-related |
| 14 | c2apoA_
|
|
 |
53.9 |
16 |
PDB header:isomerase/rna binding protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
|
| 15 | c3uaiA_
|
|
 |
48.5 |
10 |
PDB header:isomerase/chaperone
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 4;
PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
|
| 16 | c2ey4A_
|
|
 |
43.6 |
13 |
PDB header:isomerase/biosynthetic protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
|
| 17 | d1yh5a1
|
|
 |
38.7 |
19 |
Fold:YggU-like
Superfamily:YggU-like
Family:YggU-like |
| 18 | d1llda2
|
|
 |
33.0 |
9 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 19 | c2dceA_
|
|
 |
31.2 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:kiaa1915 protein;
PDBTitle: solution structure of the swirm domain of human kiaa19152 protein
|
| 20 | d2j85a1
|
|
 |
30.7 |
13 |
Fold:STIV B116-like
Superfamily:STIV B116-like
Family:STIV B116-like |
| 21 | d1mlda2 |
|
not modelled |
26.9 |
13 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 22 | c2km1A_ |
|
not modelled |
26.6 |
19 |
PDB header:protein binding
Chain: A: PDB Molecule:protein dre2;
PDBTitle: solution structure of the n-terminal domain of the yeast protein dre2
|
| 23 | d5mdha2 |
|
not modelled |
26.3 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 24 | d1utaa_ |
|
not modelled |
26.2 |
15 |
Fold:Ferredoxin-like
Superfamily:Sporulation related repeat
Family:Sporulation related repeat |
| 25 | d1t2da2 |
|
not modelled |
26.2 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 26 | d2j6ba1 |
|
not modelled |
26.0 |
14 |
Fold:STIV B116-like
Superfamily:STIV B116-like
Family:STIV B116-like |
| 27 | d1oc4a2 |
|
not modelled |
25.8 |
18 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 28 | c3kopB_ |
|
not modelled |
25.5 |
29 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
|
| 29 | d1ez4a2 |
|
not modelled |
25.4 |
22 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 30 | d1b8pa2 |
|
not modelled |
25.2 |
5 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 31 | d1hyha2 |
|
not modelled |
25.0 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 32 | c2v9vA_ |
|
not modelled |
24.9 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: crystal structure of moorella thermoacetica selb(377-511)
|
| 33 | d7mdha2 |
|
not modelled |
24.7 |
18 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 34 | c2kvrA_ |
|
not modelled |
24.6 |
5 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin carboxyl-terminal hydrolase 7;
PDBTitle: solution nmr structure of human ubiquitin specific protease2 usp7 ubl domain (residues 537-664). nesg target hr4395c/3 sgc-toronto
|
| 35 | d1gv0a2 |
|
not modelled |
24.6 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 36 | c3hskB_ |
|
not modelled |
24.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 with nadp from candida albicans
|
| 37 | d1obba2 |
|
not modelled |
24.2 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:AglA-like glucosidase |
| 38 | d9ldta2 |
|
not modelled |
24.2 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 39 | d2cmda2 |
|
not modelled |
24.0 |
13 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 40 | d1guza2 |
|
not modelled |
22.9 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 41 | d1o6za2 |
|
not modelled |
22.4 |
9 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 42 | c3f1xA_ |
|
not modelled |
21.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
| 43 | d1uxja2 |
|
not modelled |
21.3 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 44 | d1pzga2 |
|
not modelled |
21.2 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 45 | d1civa2 |
|
not modelled |
21.1 |
18 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 46 | d1hyea2 |
|
not modelled |
21.1 |
32 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 47 | c3mpoD_ |
|
not modelled |
21.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted hydrolase of the had superfamily;
PDBTitle: the crystal structure of a hydrolase from lactobacillus brevis
|
| 48 | d1y7ta2 |
|
not modelled |
21.0 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 49 | d1i0za2 |
|
not modelled |
21.0 |
17 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 50 | d1ldma2 |
|
not modelled |
20.9 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 51 | d2dw4a1 |
|
not modelled |
20.8 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:SWIRM domain |
| 52 | c3lhoA_ |
|
not modelled |
20.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative hydrolase (yp_751971.1) from shewanella2 frigidimarina ncimb 400 at 1.80 a resolution
|
| 53 | d1zpwx1 |
|
not modelled |
20.8 |
10 |
Fold:Ferredoxin-like
Superfamily:TTP0101/SSO1404-like
Family:TTP0101/SSO1404-like |
| 54 | d1w4xa1 |
|
not modelled |
20.6 |
67 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 55 | d2nn4a1 |
|
not modelled |
20.1 |
25 |
Fold:YqgQ-like
Superfamily:YqgQ-like
Family:YqgQ-like |
| 56 | d1ldna2 |
|
not modelled |
19.7 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 57 | d1a5za2 |
|
not modelled |
19.7 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 58 | d1llca2 |
|
not modelled |
19.7 |
21 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 59 | d1i10a2 |
|
not modelled |
19.1 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 60 | d2pg4a1 |
|
not modelled |
19.0 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:F93-like |
| 61 | c3cjlA_ |
|
not modelled |
18.6 |
24 |
PDB header:unknown function
Chain: A: PDB Molecule:domain of unknown function;
PDBTitle: crystal structure of a protein of unknown function (eca1910) from2 pectobacterium atrosepticum scri1043 at 2.20 a resolution
|
| 62 | d2fq3a1 |
|
not modelled |
17.8 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:SWIRM domain |
| 63 | d2dkya1 |
|
not modelled |
17.3 |
18 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Variant SAM domain |
| 64 | d2g17a1 |
|
not modelled |
17.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 65 | d1vkna1 |
|
not modelled |
16.9 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 66 | d2h80a1 |
|
not modelled |
16.6 |
18 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Variant SAM domain |
| 67 | c2hjsA_ |
|
not modelled |
16.2 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usg-1 protein homolog;
PDBTitle: the structure of a probable aspartate-semialdehyde dehydrogenase from2 pseudomonas aeruginosa
|
| 68 | d2v9va2 |
|
not modelled |
16.1 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 69 | d2hjsa1 |
|
not modelled |
16.0 |
0 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 70 | c2zv3E_ |
|
not modelled |
15.6 |
13 |
PDB header:hydrolase
Chain: E: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
|
| 71 | d2ox7a1 |
|
not modelled |
15.4 |
45 |
Fold:YopX-like
Superfamily:YopX-like
Family:YopX-like |
| 72 | d2ldxa2 |
|
not modelled |
15.4 |
5 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 73 | c3kubA_ |
|
not modelled |
15.2 |
0 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semi-aldehyde dehydrogenase complexed2 with glycerol and phosphate of mycobacterium tuberculosis h37rv
|
| 74 | c1mb4B_ |
|
not modelled |
14.9 |
0 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 from vibrio cholerae with nadp and s-methyl-l-cysteine3 sulfoxide
|
| 75 | d1y6ja2 |
|
not modelled |
14.7 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 76 | d2q49a1 |
|
not modelled |
14.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 77 | c3h92A_ |
|
not modelled |
14.3 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized atp-binding protein mjecl15;
PDBTitle: the crystal structure of one domain of the protein with unknown2 function from methanocaldococcus jannaschii
|
| 78 | c2x4iA_ |
|
not modelled |
13.9 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein 114;
PDBTitle: orf 114a from sulfolobus islandicus rudivirus 1
|
| 79 | d2cvoa1 |
|
not modelled |
13.8 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 80 | c3nwiC_ |
|
not modelled |
13.3 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:zinc transport protein zntb;
PDBTitle: the soluble domain structure of the zntb zn2+ efflux system
|
| 81 | d1qlma_ |
|
not modelled |
13.2 |
8 |
Fold:Methenyltetrahydromethanopterin cyclohydrolase
Superfamily:Methenyltetrahydromethanopterin cyclohydrolase
Family:Methenyltetrahydromethanopterin cyclohydrolase |
| 82 | c3t76A_ |
|
not modelled |
13.2 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator vanug;
PDBTitle: crystal structure of transcriptional regulator vanug, form ii
|
| 83 | d2dt5a1 |
|
not modelled |
13.1 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional repressor Rex, N-terminal domain |
| 84 | c1t4bB_ |
|
not modelled |
12.9 |
0 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: 1.6 angstrom structure of esherichia coli aspartate-2 semialdehyde dehydrogenase.
|
| 85 | d2p84a1 |
|
not modelled |
12.6 |
33 |
Fold:YopX-like
Superfamily:YopX-like
Family:YopX-like |
| 86 | d2hjja1 |
|
not modelled |
12.2 |
33 |
Fold:dsRBD-like
Superfamily:YcfA/nrd intein domain
Family:YkfF-like |
| 87 | c2hjjA_ |
|
not modelled |
12.2 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ykff;
PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
|
| 88 | c3ofeB_ |
|
not modelled |
12.1 |
11 |
PDB header:chaperone
Chain: B: PDB Molecule:ldlr chaperone boca;
PDBTitle: structured domain of drosophila melanogaster boca p41 2 2 crystal form
|
| 89 | c1jegB_ |
|
not modelled |
12.1 |
67 |
PDB header:transferase/hydrolase
Chain: B: PDB Molecule:hematopoietic cell protein-tyrosine phosphatase
PDBTitle: solution structure of the sh3 domain from c-terminal src2 kinase complexed with a peptide from the tyrosine3 phosphatase pep
|
| 90 | d1xrsb2 |
|
not modelled |
12.0 |
44 |
Fold:Dodecin subunit-like
Superfamily:D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain
Family:D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain |
| 91 | c2gz3D_ |
|
not modelled |
11.8 |
0 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aspartate beta-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate semialdehyde dehydrogenase (asadh) from2 streptococcus pneumoniae complexed with nadp and aspartate-3 semialdehyde
|
| 92 | c2qyzA_ |
|
not modelled |
11.6 |
36 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the uncharacterized protein ctc02137 from2 clostridium tetani e88
|
| 93 | d1up7a2 |
|
not modelled |
11.5 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:AglA-like glucosidase |
| 94 | d1s6ya2 |
|
not modelled |
11.5 |
38 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:AglA-like glucosidase |
| 95 | c3uw3A_ |
|
not modelled |
10.7 |
0 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of an aspartate-semialdehyde dehydrogenase from2 burkholderia thailandensis
|
| 96 | c2wh7A_ |
|
not modelled |
10.5 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronidase-phage associated;
PDBTitle: the partial structure of a group a streptpcoccal phage-2 encoded tail fibre hyaluronate lyase hylp2
|
| 97 | c2i9sA_ |
|
not modelled |
9.9 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:mesoderm development candidate 2;
PDBTitle: the solution structure of the core of mesoderm development2 (mesd).
|
| 98 | c2w3pB_ |
|
not modelled |
9.6 |
36 |
PDB header:lyase
Chain: B: PDB Molecule:benzoyl-coa-dihydrodiol lyase;
PDBTitle: boxc crystal structure
|
| 99 | c3gwdA_ |
|
not modelled |
9.3 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cyclohexanone monooxygenase;
PDBTitle: closed crystal structure of cyclohexanone monooxygenase
|