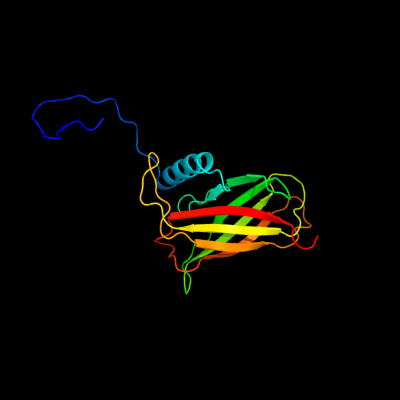
| 1 | c2jwyA_
|
|
 |
100.0 |
100 |
PDB header:lipoprotein
Chain: A: PDB Molecule:uncharacterized lipoprotein yaji;
PDBTitle: solution nmr structure of uncharacterized lipoprotein yaji from2 escherichia coli. northeast structural genomics target er540
|
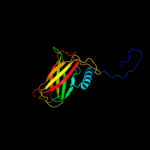
| 2 | c1kddC_
|
|
 |
83.8 |
29 |
PDB header:de novo protein
Chain: C: PDB Molecule:gcn4 acid base heterodimer acid-d12la16i;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
|
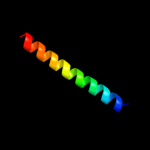
| 3 | c1kddF_
|
|
 |
83.6 |
29 |
PDB header:de novo protein
Chain: F: PDB Molecule:gcn4 acid base heterodimer acid-d12la16i;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
|
| 4 | c1kddA_
|
|
 |
83.6 |
29 |
PDB header:de novo protein
Chain: A: PDB Molecule:gcn4 acid base heterodimer acid-d12la16i;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
|
| 5 | c1kd9A_
|
|
 |
83.3 |
29 |
PDB header:de novo protein
Chain: A: PDB Molecule:gcn4 acid base heterodimer acid-d12la16l;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
|
| 6 | c1kd9C_
|
|
 |
83.3 |
29 |
PDB header:de novo protein
Chain: C: PDB Molecule:gcn4 acid base heterodimer acid-d12la16l;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
|
| 7 | c1kd9F_
|
|
 |
83.3 |
29 |
PDB header:de novo protein
Chain: F: PDB Molecule:gcn4 acid base heterodimer acid-d12la16l;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
|
| 8 | c1kd8F_
|
|
 |
69.2 |
26 |
PDB header:de novo protein
Chain: F: PDB Molecule:gcn4 acid base heterodimer acid-d12ia16v;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
|
| 9 | c1kd8C_
|
|
 |
69.2 |
26 |
PDB header:de novo protein
Chain: C: PDB Molecule:gcn4 acid base heterodimer acid-d12ia16v;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
|
| 10 | c1kd8A_
|
|
 |
67.8 |
26 |
PDB header:de novo protein
Chain: A: PDB Molecule:gcn4 acid base heterodimer acid-d12ia16v;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
|
| 11 | c2x7aB_
|
|
 |
66.1 |
20 |
PDB header:immune system
Chain: B: PDB Molecule:bone marrow stromal antigen 2;
PDBTitle: structural basis of hiv-1 tethering to membranes by the2 bst2-tetherin ectodomain
|
| 12 | c3mkxC_
|
|
 |
56.6 |
20 |
PDB header:antiviral protein
Chain: C: PDB Molecule:bone marrow stromal antigen 2;
PDBTitle: crystal structure of bst2/tetherin
|
| 13 | c2zzjA_
|
|
 |
52.2 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:glucuronan lyase a;
PDBTitle: crystal structure of endo-beta-1,4-glucuronan lyase from2 fungus trichoderma reesei
|
| 14 | c1dipA_
|
|
 |
42.9 |
24 |
PDB header:acetylation
Chain: A: PDB Molecule:delta-sleep-inducing peptide immunoreactive
PDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
|
| 15 | c1yycA_
|
|
 |
40.4 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative late embryogenesis abundant protein;
PDBTitle: solution structure of a putative late embryogenesis2 abundant (lea) protein at2g46140.1
|
| 16 | c2gr7C_
|
|
 |
38.0 |
19 |
PDB header:membrane protein
Chain: C: PDB Molecule:adhesin;
PDBTitle: hia 992-1098
|
| 17 | d2gr7a1
|
|
 |
38.0 |
19 |
Fold:Pili subunits
Superfamily:Pili subunits
Family:YadA C-terminal domain-like |
| 18 | c1u2uA_
|
|
 |
37.8 |
37 |
PDB header:transcription
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: nmr solution structure of a designed heterodimeric leucine2 zipper
|
| 19 | c1kd8E_
|
|
 |
37.1 |
23 |
PDB header:de novo protein
Chain: E: PDB Molecule:gcn4 acid base heterodimer base-d12la16l;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
|
| 20 | c3bvhC_
|
|
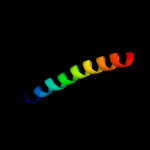 |
34.5 |
20 |
PDB header:blood clotting
Chain: C: PDB Molecule:fibrinogen gamma chain;
PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
|
| 21 | c3bkpA_ |
|
not modelled |
33.3 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:cyclophilin;
PDBTitle: crystal structure of the toxoplasma gondii cyclophilin, 49.m03261
|
| 22 | c2yy0D_ |
|
not modelled |
32.7 |
21 |
PDB header:transcription
Chain: D: PDB Molecule:c-myc-binding protein;
PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
|
| 23 | d1xo8a_ |
|
not modelled |
29.4 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:LEA14-like
Family:LEA14-like |
| 24 | c2hpcF_ |
|
not modelled |
26.0 |
19 |
PDB header:blood clotting
Chain: F: PDB Molecule:fibrinogen, gamma polypeptide;
PDBTitle: crystal structure of fragment d from human fibrinogen complexed with2 gly-pro-arg-pro-amide.
|
| 25 | d1r8ia_ |
|
not modelled |
25.6 |
11 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Typo IV secretion system protein TraC
Family:Typo IV secretion system protein TraC |
| 26 | c1r8iA_ |
|
not modelled |
25.6 |
11 |
PDB header:structural protein
Chain: A: PDB Molecule:trac;
PDBTitle: crystal structure of trac
|
| 27 | c2vefB_ |
|
not modelled |
24.9 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 28 | c1aq5C_ |
|
not modelled |
24.6 |
11 |
PDB header:coiled-coil
Chain: C: PDB Molecule:cartilage matrix protein;
PDBTitle: high-resolution solution nmr structure of the trimeric coiled-coil2 domain of chicken cartilage matrix protein, 20 structures
|
| 29 | d1o75a2 |
|
not modelled |
24.5 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Tp47 lipoprotein, middle and C-terminal domains
Family:Tp47 lipoprotein, middle and C-terminal domains |
| 30 | c3t97A_ |
|
not modelled |
22.9 |
3 |
PDB header:protein transport
Chain: A: PDB Molecule:nuclear pore glycoprotein p62;
PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup62/nup54
|
| 31 | d1afwa2 |
|
not modelled |
22.6 |
21 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 32 | c2xzrA_ |
|
not modelled |
22.5 |
15 |
PDB header:cell adhesion
Chain: A: PDB Molecule:immunoglobulin-binding protein eibd;
PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
|
| 33 | c3iynR_ |
|
not modelled |
20.1 |
10 |
PDB header:virus
Chain: R: PDB Molecule:hexon-associated protein;
PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
|
| 34 | d1smoa_ |
|
not modelled |
20.1 |
18 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 35 | d1an2a_ |
|
not modelled |
19.7 |
9 |
Fold:HLH-like
Superfamily:HLH, helix-loop-helix DNA-binding domain
Family:HLH, helix-loop-helix DNA-binding domain |
| 36 | c1xnaA_ |
|
not modelled |
19.6 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein (dna-repair protein xrcc1);
PDBTitle: nmr solution structure of the single-strand break repair2 protein xrcc1-n-terminal domain
|
| 37 | d1eqwa_ |
|
not modelled |
19.2 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Cu,Zn superoxide dismutase-like
Family:Cu,Zn superoxide dismutase-like |
| 38 | c2dzaA_ |
|
not modelled |
19.1 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 39 | d1eyea_ |
|
not modelled |
19.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 40 | c1cz7C_ |
|
not modelled |
18.4 |
19 |
PDB header:contractile protein
Chain: C: PDB Molecule:microtubule motor protein ncd;
PDBTitle: the crystal structure of a minus-end directed microtubule2 motor protein ncd reveals variable dimer conformations
|
| 41 | c3aqqD_ |
|
not modelled |
18.0 |
18 |
PDB header:dna binding protein
Chain: D: PDB Molecule:calcium-regulated heat stable protein 1;
PDBTitle: crystal structure of human crhsp-24
|
| 42 | c1kzzA_ |
|
not modelled |
17.7 |
7 |
PDB header:signaling protein
Chain: A: PDB Molecule:tnf receptor associated factor 3;
PDBTitle: downstream regulator tank binds to the cd40 recognition2 site on traf3
|
| 43 | c2y5sA_ |
|
not modelled |
17.5 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 44 | c2e43A_ |
|
not modelled |
17.1 |
15 |
PDB header:transcription/dna
Chain: A: PDB Molecule:ccaat/enhancer-binding protein beta;
PDBTitle: crystal structure of c/ebpbeta bzip homodimer k269a mutant2 bound to a high affinity dna fragment
|
| 45 | d1wu9a1 |
|
not modelled |
16.9 |
14 |
Fold:EB1 dimerisation domain-like
Superfamily:EB1 dimerisation domain-like
Family:EB1 dimerisation domain-like |
| 46 | c2ibyD_ |
|
not modelled |
16.9 |
5 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coa acetyltransferase;
PDBTitle: crystallographic and kinetic studies of human mitochondrial2 acetoacetyl-coa thiolase (t2): the importance of potassium and3 chloride for its structure and function
|
| 47 | c2bmbA_ |
|
not modelled |
16.8 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 48 | c1coiA_ |
|
not modelled |
16.2 |
19 |
PDB header:alpha-helical bundle
Chain: A: PDB Molecule:coil-vald;
PDBTitle: designed trimeric coiled coil-vald
|
| 49 | c2xv5A_ |
|
not modelled |
16.1 |
7 |
PDB header:structural protein
Chain: A: PDB Molecule:lamin-a/c;
PDBTitle: human lamin a coil 2b fragment
|
| 50 | c1n73C_ |
|
not modelled |
15.9 |
13 |
PDB header:blood clotting
Chain: C: PDB Molecule:fibrin gamma chain;
PDBTitle: fibrin d-dimer, lamprey complexed with the peptide ligand: gly-his-2 arg-pro-amide
|
| 51 | c1deqF_ |
|
not modelled |
15.5 |
16 |
PDB header:
PDB COMPND:
|
| 52 | d1e5ma2 |
|
not modelled |
15.5 |
14 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 53 | c2vp8A_ |
|
not modelled |
15.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 54 | c3n4xB_ |
|
not modelled |
15.3 |
14 |
PDB header:replication
Chain: B: PDB Molecule:monopolin complex subunit csm1;
PDBTitle: structure of csm1 full-length
|
| 55 | c1deqO_ |
|
not modelled |
14.9 |
13 |
PDB header:
PDB COMPND:
|
| 56 | d1esoa_ |
|
not modelled |
14.7 |
30 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Cu,Zn superoxide dismutase-like
Family:Cu,Zn superoxide dismutase-like |
| 57 | c1u0iA_ |
|
not modelled |
14.5 |
20 |
PDB header:de novo protein
Chain: A: PDB Molecule:iaal-e3;
PDBTitle: iaal-e3/k3 heterodimer
|
| 58 | d1v5ja_ |
|
not modelled |
14.4 |
7 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 59 | d1ajza_ |
|
not modelled |
14.4 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 60 | d1ad1a_ |
|
not modelled |
14.4 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 61 | c3pp5A_ |
|
not modelled |
14.1 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:brk1;
PDBTitle: high-resolution structure of the trimeric scar/wave complex precursor2 brk1
|
| 62 | d1m3ka2 |
|
not modelled |
13.9 |
20 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 63 | c3mcnA_ |
|
not modelled |
13.7 |
43 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
| 64 | c3ghgK_ |
|
not modelled |
13.6 |
3 |
PDB header:blood clotting
Chain: K: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of human fibrinogen
|
| 65 | d2ix4a2 |
|
not modelled |
13.6 |
29 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 66 | c1txqB_ |
|
not modelled |
13.5 |
14 |
PDB header:structural protein/protein binding
Chain: B: PDB Molecule:microtubule-associated protein rp/eb family
PDBTitle: crystal structure of the eb1 c-terminal domain complexed2 with the cap-gly domain of p150glued
|
| 67 | d1txqb1 |
|
not modelled |
13.5 |
14 |
Fold:EB1 dimerisation domain-like
Superfamily:EB1 dimerisation domain-like
Family:EB1 dimerisation domain-like |
| 68 | c3lssA_ |
|
not modelled |
13.4 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:seryl-trna synthetase;
PDBTitle: trypanosoma brucei seryl-trna synthetase in complex with atp
|
| 69 | d2gfva2 |
|
not modelled |
13.3 |
43 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 70 | c1r48A_ |
|
not modelled |
13.2 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:proline/betaine transporter;
PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
|
| 71 | d1ulqa2 |
|
not modelled |
13.0 |
27 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 72 | d1k32a4 |
|
not modelled |
12.9 |
31 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Tail specific protease, catalytic domain |
| 73 | c3p8cE_ |
|
not modelled |
12.9 |
25 |
PDB header:protein binding
Chain: E: PDB Molecule:probable protein brick1;
PDBTitle: structure and control of the actin regulatory wave complex
|
| 74 | c3cm9J_ |
|
not modelled |
12.9 |
25 |
PDB header:immune system
Chain: J: PDB Molecule:secretory component;
PDBTitle: solution structure of human siga2
|
| 75 | c3chnJ_ |
|
not modelled |
12.9 |
25 |
PDB header:immune system
Chain: J: PDB Molecule:secretory component;
PDBTitle: solution structure of human secretory iga1
|
| 76 | c3rbgB_ |
|
not modelled |
12.7 |
17 |
PDB header:immune system
Chain: B: PDB Molecule:cytotoxic and regulatory t-cell molecule;
PDBTitle: crystal structure analysis of class-i mhc restricted t-cell associated2 molecule
|
| 77 | c1tx2A_ |
|
not modelled |
12.7 |
42 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 78 | d1tx2a_ |
|
not modelled |
12.7 |
42 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 79 | d1wdkc2 |
|
not modelled |
12.6 |
13 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 80 | d2ijra1 |
|
not modelled |
12.4 |
11 |
Fold:Api92-like
Superfamily:Api92-like
Family:Api92-like |
| 81 | d1ox0a2 |
|
not modelled |
12.3 |
36 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 82 | c3ni0A_ |
|
not modelled |
12.2 |
32 |
PDB header:immune system
Chain: A: PDB Molecule:bone marrow stromal antigen 2;
PDBTitle: crystal structure of mouse bst-2/tetherin ectodomain
|
| 83 | c3tr9A_ |
|
not modelled |
12.2 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 84 | d1hkfa_ |
|
not modelled |
12.1 |
8 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 85 | c2xdjF_ |
|
not modelled |
12.1 |
6 |
PDB header:unknown function
Chain: F: PDB Molecule:uncharacterized protein ybgf;
PDBTitle: crystal structure of the n-terminal domain of e.coli ybgf
|
| 86 | c1ci6A_ |
|
not modelled |
12.0 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:transcription factor atf-4;
PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
|
| 87 | c1ca9D_ |
|
not modelled |
11.9 |
23 |
PDB header:tnf signaling
Chain: D: PDB Molecule:protein (tnf receptor associated factor 2);
PDBTitle: structure of tnf receptor associated factor 2 in complex2 with a peptide from tnf-r2
|
| 88 | c1ei3E_ |
|
not modelled |
11.8 |
3 |
PDB header:
PDB COMPND:
|
| 89 | c1fmhA_ |
|
not modelled |
11.7 |
31 |
PDB header:transcription
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: nmr solution structure of a designed heterodimeric leucine2 zipper
|
| 90 | c1t2kD_ |
|
not modelled |
11.7 |
16 |
PDB header:transcription/dna
Chain: D: PDB Molecule:cyclic-amp-dependent transcription factor atf-2;
PDBTitle: structure of the dna binding domains of irf3, atf-2 and jun2 bound to dna
|
| 91 | d1nldl1 |
|
not modelled |
11.5 |
7 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 92 | c3m9bK_ |
|
not modelled |
11.5 |
15 |
PDB header:chaperone
Chain: K: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
|
| 93 | c1fosF_ |
|
not modelled |
11.3 |
16 |
PDB header:transcription/dna
Chain: F: PDB Molecule:c-jun proto-oncogene protein;
PDBTitle: two human c-fos:c-jun:dna complexes
|
| 94 | d1ub5a1 |
|
not modelled |
11.1 |
4 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 95 | c1jccC_ |
|
not modelled |
10.8 |
3 |
PDB header:membrane protein
Chain: C: PDB Molecule:major outer membrane lipoprotein;
PDBTitle: crystal structure of a novel alanine-zipper trimer at 1.7 a2 resolution, v13a,l16a,v20a,l23a,v27a,m30a,v34a mutations
|
| 96 | d1dpta_ |
|
not modelled |
10.6 |
13 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
| 97 | c1fosE_ |
|
not modelled |
10.6 |
12 |
PDB header:transcription/dna
Chain: E: PDB Molecule:p55-c-fos proto-oncogene protein;
PDBTitle: two human c-fos:c-jun:dna complexes
|
| 98 | d1h3pl1 |
|
not modelled |
10.6 |
2 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 99 | c2q7cC_ |
|
not modelled |
10.3 |
11 |
PDB header:viral protein
Chain: C: PDB Molecule:fusion protein between yeast variant gcn4 and
PDBTitle: crystal structure of iqn17
|