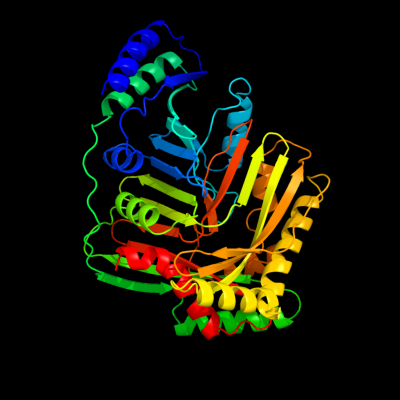
| 1 | d3bzna1
|
|
 |
100.0 |
100 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
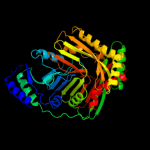
| 2 | c3gseA_
|
|
 |
100.0 |
53 |
PDB header:isomerase
Chain: A: PDB Molecule:menaquinone-specific isochorismate synthase;
PDBTitle: crystal structure of menaquinone-specific isochorismate synthase from2 yersinia pestis co92
|
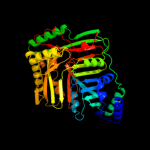
| 3 | c3hwoB_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:isochorismate synthase entc;
PDBTitle: crystal structure of escherichia coli enterobactin-specific2 isochorismate synthase entc in complex with isochorismate
|
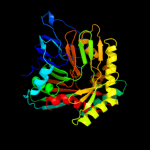
| 4 | d2g5fa1
|
|
 |
100.0 |
23 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 5 | d1qdla_
|
|
 |
100.0 |
23 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 6 | c3os6A_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:isochorismate synthase dhbc;
PDBTitle: crystal structure of putative 2,3-dihydroxybenzoate-specific2 isochorismate synthase, dhbc from bacillus anthracis.
|
| 7 | d2fn0a1
|
|
 |
100.0 |
25 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 8 | d1i1qa_
|
|
 |
100.0 |
21 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 9 | d1i7qa_
|
|
 |
100.0 |
22 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 10 | c2i6yA_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:anthranilate synthase component i, putative;
PDBTitle: structure and mechanism of mycobacterium tuberculosis salicylate2 synthase, mbti
|
| 11 | c3h9mA_
|
|
 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:p-aminobenzoate synthetase, component i;
PDBTitle: crystal structure of para-aminobenzoate synthetase,2 component i from cytophaga hutchinsonii
|
| 12 | c3r74B_
|
|
 |
100.0 |
19 |
PDB header:lyase, biosynthetic protein
Chain: B: PDB Molecule:anthranilate/para-aminobenzoate synthases component i;
PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
|
| 13 | d1k0ga_
|
|
 |
100.0 |
19 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
| 14 | c3nqkA_
|
|
 |
30.8 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a structural genomics, unknown function2 (bacova_03322) from bacteroides ovatus at 2.61 a resolution
|
| 15 | d1eyba_
|
|
 |
19.2 |
11 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Homogentisate dioxygenase |
| 16 | c1ey2A_
|
|
 |
19.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homogentisate 1,2-dioxygenase;
PDBTitle: human homogentisate dioxygenase with fe(ii)
|
| 17 | d1hxra_
|
|
 |
18.7 |
29 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
| 18 | d2fu5a1
|
|
 |
17.9 |
29 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
| 19 | d2ffca1
|
|
 |
17.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 20 | c3fuyC_
|
|
 |
14.2 |
36 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative integron gene cassette protein;
PDBTitle: structure from the mobile metagenome of cole harbour salt2 marsh: integron cassette protein hfx_cass1
|
| 21 | d2nwua1 |
|
not modelled |
12.7 |
12 |
Fold:RL5-like
Superfamily:RL5-like
Family:SSO1042-like |
| 22 | d1d5ta2 |
|
not modelled |
11.6 |
36 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GDI-like |
| 23 | c3ci9B_ |
|
not modelled |
10.3 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:heat shock factor-binding protein 1;
PDBTitle: crystal structure of the human hsbp1
|
| 24 | d1ji8a_ |
|
not modelled |
9.7 |
26 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
| 25 | c1wxnA_ |
|
not modelled |
9.2 |
46 |
PDB header:toxin
Chain: A: PDB Molecule:toxin apetx2;
PDBTitle: solution structure of apetx2, a specific peptide inhibitor2 of asic3 proton-gated channels
|
| 26 | c2km1A_ |
|
not modelled |
8.7 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:protein dre2;
PDBTitle: solution structure of the n-terminal domain of the yeast protein dre2
|
| 27 | c3c66B_ |
|
not modelled |
8.7 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) polymerase;
PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
|
| 28 | d1aopa2 |
|
not modelled |
8.7 |
13 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 29 | c3ol4B_ |
|
not modelled |
8.6 |
38 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
|
| 30 | d2ch5a2 |
|
not modelled |
8.5 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 31 | c2z1tA_ |
|
not modelled |
7.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hydrogenase maturation protein hype
|
| 32 | c1wqkA_ |
|
not modelled |
7.9 |
54 |
PDB header:toxin
Chain: A: PDB Molecule:toxin apetx1;
PDBTitle: solution structure of apetx1, a specific peptide inhibitor2 of human ether-a-go-go-related gene potassium channels3 from the venom of the sea anemone anthopleura4 elegantissima: a new fold for an herg toxin
|
| 33 | c2zkrf_ |
|
not modelled |
7.4 |
20 |
PDB header:ribosomal protein/rna
Chain: F: PDB Molecule:rna expansion segment es7 part iii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 34 | c3rpdB_ |
|
not modelled |
7.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:methionine synthase (b12-independent);
PDBTitle: the structure of a b12-independent methionine synthase from shewanella2 sp. w3-18-1 in complex with selenomethionine.
|
| 35 | c1s1iG_ |
|
not modelled |
6.9 |
16 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l8-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 36 | d2bcgg3 |
|
not modelled |
6.8 |
21 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GDI-like |
| 37 | c3izcH_ |
|
not modelled |
6.1 |
16 |
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein rpl8 (l7ae);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 38 | c3rrrB_ |
|
not modelled |
6.0 |
45 |
PDB header:viral protein
Chain: B: PDB Molecule:fusion glycoprotein f0;
PDBTitle: structure of the rsv f protein in the post-fusion conformation
|
| 39 | c3t6mA_ |
|
not modelled |
6.0 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:succinyl-diaminopimelate desuccinylase;
PDBTitle: crystal structure of the catalytic domain of dape protein from2 v.cholerea in the zn bound form
|
| 40 | c4a1eF_ |
|
not modelled |
5.7 |
22 |
PDB header:ribosome
Chain: F: PDB Molecule:rpl7a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
|
| 41 | d2c1wa1 |
|
not modelled |
5.6 |
50 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Eukaryotic EndoU ribonuclease |
| 42 | c3dwlG_ |
|
not modelled |
5.6 |
14 |
PDB header:structural protein
Chain: G: PDB Molecule:actin-related protein 2/3 complex subunit 5;
PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
|
| 43 | c3fhdA_ |
|
not modelled |
5.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:orf 37;
PDBTitle: crystal structure of the shutoff and exonuclease protein from kaposis2 sarcoma associated herpesvirus
|
| 44 | d2fkia1 |
|
not modelled |
5.5 |
14 |
Fold:Secretion chaperone-like
Superfamily:YjbR-like
Family:YjbR-like |
| 45 | d1ydua1 |
|
not modelled |
5.1 |
7 |
Fold:At5g01610-like
Superfamily:At5g01610-like
Family:At5g01610-like |
| 46 | c2kvcA_ |
|
not modelled |
5.1 |
24 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
|