| 1 |
|
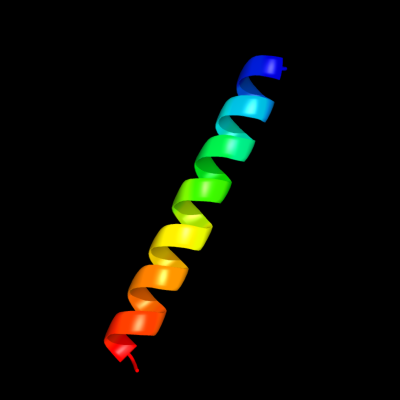
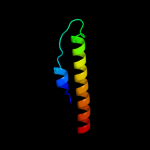
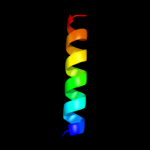
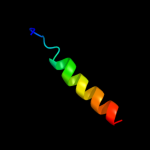
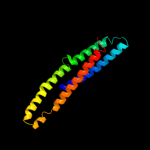
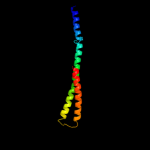
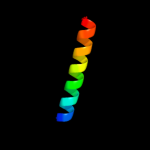
PDB 2osz chain A
Region: 145 - 171
Aligned: 27
Modelled: 27
Confidence: 57.0%
Identity: 33%
PDB header:structural protein
Chain: A: PDB Molecule:nucleoporin p58/p45;
PDBTitle: structure of nup58/45 suggests flexible nuclear pore diameter by2 intermolecular sliding
Phyre2
| 2 |
|
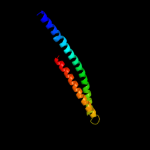
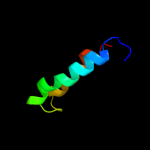
PDB 1ei3 chain C
Region: 52 - 170
Aligned: 112
Modelled: 114
Confidence: 27.9%
Identity: 10%
PDB header:
PDB COMPND:
Phyre2
| 3 |
|
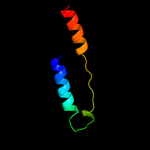
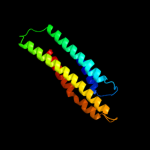
PDB 1deq chain F
Region: 48 - 170
Aligned: 109
Modelled: 123
Confidence: 17.6%
Identity: 9%
PDB header:
PDB COMPND:
Phyre2
| 4 |
|
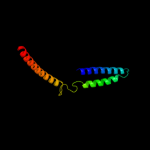
PDB 3oja chain B
Region: 40 - 174
Aligned: 130
Modelled: 130
Confidence: 17.0%
Identity: 16%
PDB header:protein binding
Chain: B: PDB Molecule:anopheles plasmodium-responsive leucine-rich repeat protein
PDBTitle: crystal structure of lrim1/apl1c complex
Phyre2
| 5 |
|
PDB 3u59 chain C
Region: 77 - 174
Aligned: 95
Modelled: 98
Confidence: 15.3%
Identity: 7%
PDB header:contractile protein
Chain: C: PDB Molecule:tropomyosin beta chain;
PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
Phyre2
| 6 |
|
PDB 1deq chain O
Region: 110 - 174
Aligned: 63
Modelled: 65
Confidence: 14.4%
Identity: 11%
PDB header:
PDB COMPND:
Phyre2
| 7 |
|
PDB 2wuk chain D
Region: 23 - 70
Aligned: 48
Modelled: 48
Confidence: 14.3%
Identity: 6%
PDB header:cell cycle
Chain: D: PDB Molecule:septum site-determining protein diviva;
PDBTitle: diviva n-terminal domain, f17a mutant
Phyre2
| 8 |
|
PDB 2xus chain A
Region: 153 - 173
Aligned: 21
Modelled: 21
Confidence: 13.5%
Identity: 24%
PDB header:protein binding
Chain: A: PDB Molecule:breast cancer metastasis-suppressor 1;
PDBTitle: crystal structure of the brms1 n-terminal region
Phyre2
| 9 |
|
PDB 1k1f chain A
Region: 149 - 172
Aligned: 24
Modelled: 24
Confidence: 13.0%
Identity: 21%
Fold: Bcr-Abl oncoprotein oligomerization domain
Superfamily: Bcr-Abl oncoprotein oligomerization domain
Family: Bcr-Abl oncoprotein oligomerization domain
Phyre2
| 10 |
|
PDB 3cwg chain A
Region: 41 - 175
Aligned: 129
Modelled: 135
Confidence: 10.9%
Identity: 7%
PDB header:transcription
Chain: A: PDB Molecule:signal transducer and activator of transcription
PDBTitle: unphosphorylated mouse stat3 core fragment
Phyre2
| 11 |
|
PDB 2efr chain B
Region: 7 - 175
Aligned: 151
Modelled: 154
Confidence: 10.3%
Identity: 11%
PDB header:contractile protein
Chain: B: PDB Molecule:general control protein gcn4 and tropomyosin 1 alpha chain;
PDBTitle: crystal structure of the c-terminal tropomyosin fragment with n- and2 c-terminal extensions of the leucine zipper at 1.8 angstroms3 resolution
Phyre2
| 12 |
|
PDB 3plt chain B
Region: 78 - 175
Aligned: 94
Modelled: 98
Confidence: 10.2%
Identity: 14%
PDB header:structural protein
Chain: B: PDB Molecule:sphingolipid long chain base-responsive protein lsp1;
PDBTitle: crystal structure of lsp1 from saccharomyces cerevisiae
Phyre2
| 13 |
|
PDB 2gl2 chain B
Region: 79 - 174
Aligned: 93
Modelled: 96
Confidence: 9.2%
Identity: 13%
PDB header:cell adhesion
Chain: B: PDB Molecule:adhesion a;
PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
Phyre2
| 14 |
|
PDB 2gts chain A domain 1
Region: 7 - 52
Aligned: 46
Modelled: 46
Confidence: 7.7%
Identity: 24%
Fold: Ferritin-like
Superfamily: HP0062-like
Family: HP0062-like
Phyre2
| 15 |
|
PDB 1ykh chain B domain 1
Region: 40 - 139
Aligned: 96
Modelled: 100
Confidence: 6.2%
Identity: 13%
Fold: Mediator hinge subcomplex-like
Superfamily: Mediator hinge subcomplex-like
Family: CSE2-like
Phyre2
| 16 |
|
PDB 2x6p chain B
Region: 143 - 167
Aligned: 25
Modelled: 25
Confidence: 5.9%
Identity: 32%
PDB header:de novo protein
Chain: B: PDB Molecule:coil ser l19c;
PDBTitle: crystal structure of coil ser l19c
Phyre2
| 17 |
|
PDB 2x6p chain A
Region: 143 - 167
Aligned: 25
Modelled: 25
Confidence: 5.9%
Identity: 32%
PDB header:de novo protein
Chain: A: PDB Molecule:coil ser l19c;
PDBTitle: crystal structure of coil ser l19c
Phyre2
| 18 |
|
PDB 1sdi chain A
Region: 1 - 49
Aligned: 46
Modelled: 46
Confidence: 5.7%
Identity: 9%
Fold: YcfC-like
Superfamily: YcfC-like
Family: YcfC-like
Phyre2
| 19 |
|
PDB 2x6p chain C
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 5.5%
Identity: 31%
PDB header:de novo protein
Chain: C: PDB Molecule:coil ser l19c;
PDBTitle: crystal structure of coil ser l19c
Phyre2
| 20 |
|
PDB 1gs9 chain A
Region: 5 - 134
Aligned: 129
Modelled: 130
Confidence: 5.3%
Identity: 15%
Fold: Four-helical up-and-down bundle
Superfamily: Apolipoprotein
Family: Apolipoprotein
Phyre2