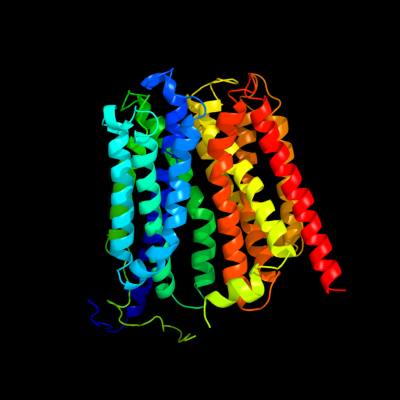
1 d1pw4a_
100.0
12
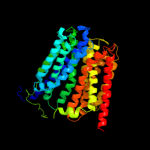
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter2 c3o7pA_
100.0
12
PDB header: transport proteinChain: A: PDB Molecule: l-fucose-proton symporter;PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
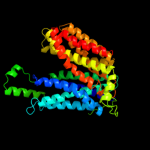
3 d1pv7a_
100.0
12
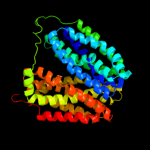
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter4 c2gfpA_
100.0
10
PDB header: membrane proteinChain: A: PDB Molecule: multidrug resistance protein d;PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
5 c2xutC_
99.9
13
PDB header: transport proteinChain: C: PDB Molecule: proton/peptide symporter family protein;PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
6 c3b9yA_
55.5
13
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
7 c2bbjB_
34.2
10
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
8 c3obhA_
26.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: x-ray crystal structure of protein sp_0782 (7-79) from streptococcus2 pneumoniae. northeast structural genomics consortium target spr104
9 c3db3A_
26.1
19
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase uhrf1;PDBTitle: crystal structure of the tandem tudor domains of the e3 ubiquitin-2 protein ligase uhrf1 in complex with trimethylated histone h3-k93 peptide
10 c3metB_
23.6
11
PDB header: transcriptionChain: B: PDB Molecule: saga-associated factor 29 homolog;PDBTitle: crystal structure of sgf29 in complex with h3k4me2
11 c2l92A_
20.6
22
PDB header: dna binding proteinChain: A: PDB Molecule: histone family protein nucleoid-structuring protein h-ns;PDBTitle: solution structure of the c-terminal domain of h-ns like protein bv3f
12 c3pm7A_
18.6
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ef_3132 protein from enterococcus faecalis at the2 resolution 2a, northeast structural genomics consortium target efr184
13 c1hw4A_
17.2
17
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase;PDBTitle: structure of thymidylate synthase suggests advantages of chemotherapy2 with noncompetitive inhibitors
14 c1hw3A_
14.6
17
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase;PDBTitle: structure of human thymidylate synthase suggests advantages of2 chemotherapy with noncompetitive inhibitors
15 c2kncA_
14.6
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
16 c3hd6A_
12.3
13
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
17 c2kz7C_
11.9
27
PDB header: protein bindingChain: C: PDB Molecule: leucine-rich repeat-containing protein 16a;PDBTitle: solution structure of the carmil cah3a/b domain bound to capping2 protein (cp)
18 d2iuba2
11.6
17
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region19 c2g9pA_
11.5
14
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide latarcin 2a;PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
20 c3dgpB_
8.4
20
PDB header: transcriptionChain: B: PDB Molecule: rna polymerase ii transcription factor b subunit 5;PDBTitle: crystal structure of the complex between tfb5 and the c-terminal2 domain of tfb2
21 c3ke2A_
not modelled
8.1
20
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yp_928783.1;PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
22 c3mkuA_
not modelled
7.9
10
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
23 d1sv0c_
not modelled
7.7
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain24 d1sxda_
not modelled
7.5
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain25 c2dkxA_
not modelled
7.2
18
PDB header: signaling proteinChain: A: PDB Molecule: sam pointed domain-containing ets transcriptionPDBTitle: solution structure of the sam_pnt-domain of ets2 transcription factor pdef (prostate ets)
26 d1hnra_
not modelled
7.1
18
Fold: H-NS histone-like proteinsSuperfamily: H-NS histone-like proteinsFamily: H-NS histone-like proteins27 d1bcoa1
not modelled
7.0
7
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain28 d1sv0a_
not modelled
6.1
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain29 c1y6gB_
not modelled
6.1
33
PDB header: transferase/dnaChain: B: PDB Molecule: dna alpha-glucosyltransferase;PDBTitle: alpha-glucosyltransferase in complex with udp and a 13_mer2 dna containing a hmu base at 2.8 a resolution
30 d1vbva1
not modelled
6.1
22
Fold: SH3-like barrelSuperfamily: YccV-likeFamily: YccV-like31 d1bqva_
not modelled
5.9
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain32 d1jb0a_
not modelled
5.9
20
Fold: Photosystem I subunits PsaA/PsaBSuperfamily: Photosystem I subunits PsaA/PsaBFamily: Photosystem I subunits PsaA/PsaB33 d2z15a1
not modelled
5.7
50
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like34 d3e9va1
not modelled
5.6
50
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like35 c2h31A_
not modelled
5.6
12
PDB header: ligase, lyaseChain: A: PDB Molecule: multifunctional protein ade2;PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
36 d1jb0b_
not modelled
5.5
16
Fold: Photosystem I subunits PsaA/PsaBSuperfamily: Photosystem I subunits PsaA/PsaBFamily: Photosystem I subunits PsaA/PsaB37 c2k9pA_
not modelled
5.5
15
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
38 c2o8kA_
not modelled
5.5
21
PDB header: transcription/dnaChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: nmr structure of the sigma-54 rpon domain bound to the-242 promoter element
39 c2yh6C_
not modelled
5.5
25
PDB header: lipid binding proteinChain: C: PDB Molecule: lipoprotein 34;PDBTitle: structure of the n-terminal domain of bamc from e. coli
40 c3hftA_
not modelled
5.5
20
PDB header: hydrolaseChain: A: PDB Molecule: wbms, polysaccharide deacetylase involved in o-antigenPDBTitle: crystal structure of a putative polysaccharide deacetylase involved in2 o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica3 at 1.90 a resolution
41 c3ehuC_
not modelled
5.4
27
PDB header: membrane proteinChain: C: PDB Molecule: corticoliberin;PDBTitle: crystal structure of the extracellular domain of human corticotropin2 releasing factor receptor type 1 (crfr1) in complex with crf
42 c3ehuD_
not modelled
5.4
27
PDB header: membrane proteinChain: D: PDB Molecule: corticoliberin;PDBTitle: crystal structure of the extracellular domain of human corticotropin2 releasing factor receptor type 1 (crfr1) in complex with crf
43 c3jyvT_
not modelled
5.4
16
PDB header: ribosomeChain: T: PDB Molecule: s19e protein;PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
44 d1sxea_
not modelled
5.2
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain45 d1hdmb2
not modelled
5.2
29
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain46 c2ytuA_
not modelled
5.1
19
PDB header: signaling proteinChain: A: PDB Molecule: friend leukemia integration 1 transcriptionPDBTitle: solution structure of the sam_pnt-domain of the human2 friend leukemiaintegration 1 transcription factor