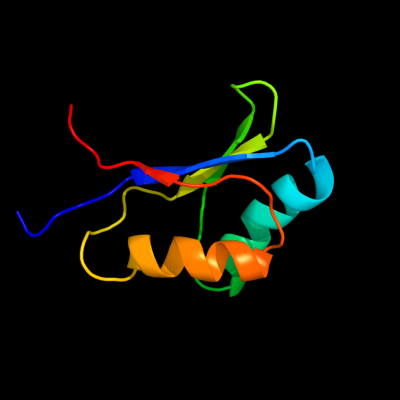
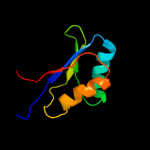
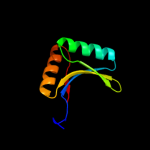
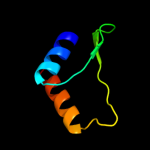
1 d1utaa_
99.9
100
Fold: Ferredoxin-likeSuperfamily: Sporulation related repeatFamily: Sporulation related repeat2 c1x60A_
99.8
29
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
3 c3caiA_
88.0
22
PDB header: transferaseChain: A: PDB Molecule: possible aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
4 d1bs0a_
87.6
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like5 d1elua_
85.5
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like6 c3f9tB_
81.6
19
PDB header: lyaseChain: B: PDB Molecule: l-tyrosine decarboxylase mfna;PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
7 d1t3ia_
65.9
23
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like8 c2k49A_
59.9
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein so_3888;PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
9 d1iuga_
59.2
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like10 c3hdoB_
59.1
24
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
11 c3cq6E_
58.3
18
PDB header: transferaseChain: E: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: histidinol-phosphate aminotransferase from corynebacterium2 glutamicum holo-form (plp covalently bound )
12 d1m32a_
58.0
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like13 c2fyfB_
56.4
18
PDB header: transferaseChain: B: PDB Molecule: phosphoserine aminotransferase;PDBTitle: structure of a putative phosphoserine aminotransferase from2 mycobacterium tuberculosis
14 c2yrrA_
52.9
23
PDB header: transferaseChain: A: PDB Molecule: aminotransferase, class v;PDBTitle: hypothetical alanine aminotransferase (tth0173) from thermus2 thermophilus hb8
15 d1qz9a_
52.8
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like16 d1v72a1
52.6
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like17 c3hqtB_
49.8
19
PDB header: transferaseChain: B: PDB Molecule: cai-1 autoinducer synthase;PDBTitle: plp-dependent acyl-coa transferase cqsa
18 d1wyua1
47.6
20
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Glycine dehydrogenase subunits (GDC-P)19 c3rggD_
46.5
19
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase, pure protein;PDBTitle: crystal structure of treponema denticola pure bound to air
20 d2f8ja1
46.5
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like21 c2k8eA_
not modelled
46.3
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein yegp;PDBTitle: solution nmr structure of protein of unknown function yegp from e.2 coli. ontario center for structural proteomics target ec0640_1_1233 northeast structural genomics consortium target et102.
22 d1eg5a_
not modelled
44.5
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like23 c3p3dA_
not modelled
44.2
21
PDB header: nuclear proteinChain: A: PDB Molecule: nucleoporin 53;PDBTitle: crystal structure of the nup53 rrm domain from pichia guilliermondii
24 d1gcca_
not modelled
43.5
26
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: GCC-box binding domain25 c2adbA_
not modelled
43.0
9
PDB header: rna binding protein/rnaChain: A: PDB Molecule: polypyrimidine tract-binding protein 1;PDBTitle: solution structure of polypyrimidine tract binding protein2 rbd2 complexed with cucucu rna
26 d1dq3a4
not modelled
41.6
14
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease27 c2y9jt_
not modelled
41.0
14
PDB header: protein transportChain: T: PDB Molecule: protein prgh;PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
28 d1u11a_
not modelled
40.0
16
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)29 c2w8wA_
not modelled
39.7
19
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: n100y spt with plp-ser
30 d2c0ra1
not modelled
38.5
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like31 c2x7iA_
not modelled
37.9
17
PDB header: transferaseChain: A: PDB Molecule: mevalonate kinase;PDBTitle: crystal structure of mevalonate kinase from methicillin-2 resistant staphylococcus aureus mrsa252
32 c3a2bA_
not modelled
37.6
13
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
33 d1ztpa1
not modelled
36.4
8
Fold: eIF4e-likeSuperfamily: eIF4e-likeFamily: BLES03-like34 d1nz8a_
not modelled
35.9
17
Fold: Ferredoxin-likeSuperfamily: N-utilization substance G protein NusG, N-terminal domainFamily: N-utilization substance G protein NusG, N-terminal domain35 d1wg1a_
not modelled
35.7
8
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD36 c2fw9A_
not modelled
35.2
16
PDB header: lyaseChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
37 d2hk6a1
not modelled
33.5
15
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase38 d1jf9a_
not modelled
33.3
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like39 c3f0hA_
not modelled
33.2
13
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
40 c3pn8A_
not modelled
32.3
18
PDB header: hydrolaseChain: A: PDB Molecule: putative phospho-beta-glucosidase;PDBTitle: the crystal structure of 6-phospho-beta-glucosidase from streptococcus2 mutans ua159
41 c3lp6D_
not modelled
32.1
21
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase catalytic subunit;PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
42 d1qcza_
not modelled
31.8
16
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)43 c3e9kA_
not modelled
31.1
17
PDB header: hydrolaseChain: A: PDB Molecule: kynureninase;PDBTitle: crystal structure of homo sapiens kynureninase-3-hydroxyhippuric acid2 inhibitor complex
44 c3trhI_
not modelled
30.8
16
PDB header: lyaseChain: I: PDB Molecule: phosphoribosylaminoimidazole carboxylasePDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
45 c2dr1A_
not modelled
29.2
11
PDB header: transferaseChain: A: PDB Molecule: 386aa long hypothetical serine aminotransferase;PDBTitle: crystal structure of the ph1308 protein from pyrococcus horikoshii ot3
46 d1m6sa_
not modelled
28.6
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like47 c1yj7A_
not modelled
28.6
12
PDB header: protein transportChain: A: PDB Molecule: escj;PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
48 d1nh8a2
not modelled
28.1
12
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain49 c3rfaA_
not modelled
27.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
50 c2k06A_
not modelled
27.5
13
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of the aminoterminal domain of e. coli2 nusg
51 c3cbfA_
not modelled
26.8
15
PDB header: transferaseChain: A: PDB Molecule: alpha-aminodipate aminotransferase;PDBTitle: crystal structure of lysn, alpha-aminoadipate2 aminotransferase, from thermus thermophilus hb27
52 c2j75A_
not modelled
26.7
18
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase a;PDBTitle: beta-glucosidase from thermotoga maritima in complex with2 noeuromycin
53 c3hyjD_
not modelled
26.5
22
PDB header: transcription regulatorChain: D: PDB Molecule: protein duf199/whia;PDBTitle: crystal structure of the n-terminal laglidadg domain of duf199/whia
54 c3orsD_
not modelled
26.3
17
PDB header: isomerase,biosynthetic proteinChain: D: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
55 d1xmpa_
not modelled
26.2
18
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)56 c3nthA_
not modelled
25.0
16
PDB header: transcriptionChain: A: PDB Molecule: maternal protein tudor;PDBTitle: crystal structure of tudor and aubergine [r13(me2s)] complex
57 c3mafB_
not modelled
24.1
17
PDB header: lyaseChain: B: PDB Molecule: sphingosine-1-phosphate lyase;PDBTitle: crystal structure of stspl (asymmetric form)
58 d1js3a_
not modelled
23.7
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Pyridoxal-dependent decarboxylase59 c2hzpA_
not modelled
23.7
17
PDB header: hydrolaseChain: A: PDB Molecule: kynureninase;PDBTitle: crystal structure of homo sapiens kynureninase
60 c2p2uA_
not modelled
23.6
41
PDB header: dna binding proteinChain: A: PDB Molecule: host-nuclease inhibitor protein gam, putative;PDBTitle: crystal structure of putative host-nuclease inhibitor2 protein gam from desulfovibrio vulgaris
61 c2h31A_
not modelled
23.3
18
PDB header: ligase, lyaseChain: A: PDB Molecule: multifunctional protein ade2;PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
62 c3nohA_
not modelled
23.1
50
PDB header: peptide binding proteinChain: A: PDB Molecule: putative peptide binding protein;PDBTitle: crystal structure of a putative peptide binding protein (rumgna_00914)2 from ruminococcus gnavus atcc 29149 at 1.60 a resolution
63 d1u0bb1
not modelled
22.5
16
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases64 c3bfjK_
not modelled
21.3
15
PDB header: oxidoreductaseChain: K: PDB Molecule: 1,3-propanediol oxidoreductase;PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
65 c2q5cA_
not modelled
21.1
18
PDB header: transcriptionChain: A: PDB Molecule: ntrc family transcriptional regulator;PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
66 d2cpia1
not modelled
21.0
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD67 d1oj4a2
not modelled
20.4
11
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE68 d1lbqa_
not modelled
20.1
13
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase69 c2huuA_
not modelled
20.1
13
PDB header: transferaseChain: A: PDB Molecule: alanine glyoxylate aminotransferase;PDBTitle: crystal structure of aedes aegypti alanine glyoxylate2 aminotransferase in complex with alanine
70 c3ffrA_
not modelled
20.0
13
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase serc;PDBTitle: crystal structure of a phosphoserine aminotransferase serc (chu_0995)2 from cytophaga hutchinsonii atcc 33406 at 1.75 a resolution
71 d1rk8a_
not modelled
19.7
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD72 d1fg7a_
not modelled
19.5
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like73 c3ox4D_
not modelled
19.3
19
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase 2;PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
74 c2eqkA_
not modelled
19.0
43
PDB header: transcriptionChain: A: PDB Molecule: tudor domain-containing protein 4;PDBTitle: solution structure of the tudor domain of tudor domain-2 containing protein 4
75 d1wsta1
not modelled
18.9
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like76 d2erra1
not modelled
18.4
8
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD77 c3o6cA_
not modelled
18.1
12
PDB header: transferaseChain: A: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
78 c3hyiA_
not modelled
18.0
18
PDB header: transcription regulatorChain: A: PDB Molecule: protein duf199/whia;PDBTitle: crystal structure of full-length duf199/whia from thermatoga maritima
79 d1s0aa_
not modelled
17.9
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like80 c2z1sA_
not modelled
17.8
18
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase b;PDBTitle: beta-glucosidase b from paenibacillus polymyxa complexed2 with cellotetraose
81 c2ordA_
not modelled
17.6
15
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
82 d1kvka2
not modelled
17.1
19
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: Mevalonate kinase83 c2hvzA_
not modelled
16.8
7
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor, arginine/serine-rich 7;PDBTitle: solution structure of the rrm domain of sr rich factor 9g8
84 c2i38A_
not modelled
16.6
12
PDB header: rna binding protein/chimeraChain: A: PDB Molecule: fusion protein consists of immunoglobin g-PDBTitle: solution structure of the rrm of srp20
85 c2zoxA_
not modelled
16.1
18
PDB header: hydrolaseChain: A: PDB Molecule: cytosolic beta-glucosidase;PDBTitle: crystal structure of the covalent intermediate of human cytosolic2 beta-glucosidase
86 c2iruA_
not modelled
16.0
24
PDB header: transferaseChain: A: PDB Molecule: putative dna ligase-like protein rv0938/mt0965;PDBTitle: crystal structure of the polymerase domain from mycobacterium2 tuberculosis ligase d
87 c2dclB_
not modelled
15.9
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical upf0166 protein ph1503;PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
88 d1fsha_
not modelled
15.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain89 c3md3A_
not modelled
15.9
4
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear and cytoplasmic polyadenylated rna-binding proteinPDBTitle: crystal structure of the first two rrm domains of yeast poly(u)2 binding protein (pub1)
90 d2p5ia1
not modelled
15.7
27
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like91 c3ml6D_
not modelled
15.5
15
PDB header: protein transportChain: D: PDB Molecule: chimeric complex between protein dishevlled2 homolog dvl-2PDBTitle: a complex between dishevlled2 and clathrin adaptor ap-2
92 d1pbga_
not modelled
15.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Family 1 of glycosyl hydrolase93 d1o51a_
not modelled
14.6
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: DUF190/COG199394 c1v02F_
not modelled
14.2
20
PDB header: hydrolaseChain: F: PDB Molecule: dhurrinase;PDBTitle: crystal structure of the sorghum bicolor dhurrinase 1
95 d1v02a_
not modelled
14.2
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Family 1 of glycosyl hydrolase96 d2a21a1
not modelled
14.2
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase97 d2k7ia1
not modelled
14.1
11
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like98 c2k7iB_
not modelled
14.1
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: upf0339 protein atu0232;PDBTitle: solution nmr structure of protein atu0232 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att3. ontario center for structural proteomics target atc0223.
99 d1rv3a_
not modelled
14.1
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like