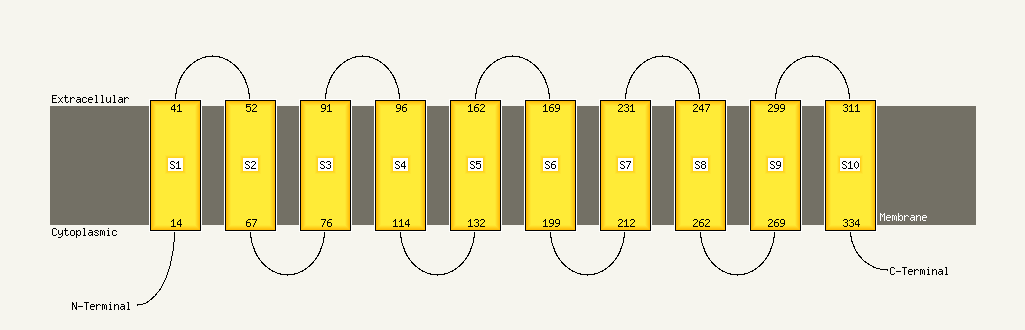
1 c3qnqD_
100.0
12
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
2 d1a3aa_
100.0
100
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: IIA domain of mannitol-specific and ntr phosphotransferase EII3 c3oxpB_
99.9
30
PDB header: transferaseChain: B: PDB Molecule: phosphotransferase enzyme ii, a component;PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
4 c3oxpA_
99.9
30
PDB header: transferaseChain: A: PDB Molecule: phosphotransferase enzyme ii, a component;PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
5 c2oq3A_
99.9
27
PDB header: transferaseChain: A: PDB Molecule: mannitol-specific cryptic phosphotransferasePDBTitle: solution structure of the mannitol- specific cryptic2 phosphotransferase enzyme iia cmtb from escherichia coli
6 c2oqtD_
99.9
26
PDB header: transferaseChain: D: PDB Molecule: hypothetical protein spy0176;PDBTitle: structural genomics, the crystal structure of a putative2 pts iia domain from streptococcus pyogenes m1 gas
7 c3bjvA_
99.9
26
PDB header: transferaseChain: A: PDB Molecule: rmpa;PDBTitle: the crystal structure of a putative pts iia(ptxa) from streptococcus2 mutans
8 c3urrB_
99.9
23
PDB header: transferaseChain: B: PDB Molecule: pts iia-like nitrogen-regulatory protein ptsn;PDBTitle: structure of pts iia-like nitrogen-regulatory protein ptsn (bth_i0484)2 (ptsn)
9 d1a6ja_
99.9
24
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: IIA domain of mannitol-specific and ntr phosphotransferase EII10 c2a0jA_
99.9
26
PDB header: transferaseChain: A: PDB Molecule: pts system, nitrogen regulatory iia protein;PDBTitle: crystal structure of nitrogen regulatory protein iia-ntr from2 neisseria meningitidis
11 d1xiza_
99.9
20
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: IIA domain of mannitol-specific and ntr phosphotransferase EII12 c1vkrA_
99.6
100
PDB header: transferaseChain: A: PDB Molecule: mannitol-specific pts system enzyme iiabc components;PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
13 d1vkra_
99.6
100
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Lactose/Cellobiose specific IIB subunit14 c3czcA_
99.4
20
PDB header: transferaseChain: A: PDB Molecule: rmpb;PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
15 c1tvmA_
99.3
20
PDB header: transferaseChain: A: PDB Molecule: pts system, galactitol-specific iib component;PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
16 c3sqnB_
99.2
9
PDB header: transcription regulatorChain: B: PDB Molecule: conserved domain protein;PDBTitle: putative mga family transcriptional regulator from enterococcus2 faecalis
17 d1iiba_
97.4
12
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Lactose/Cellobiose specific IIB subunit18 c3nbmA_
96.5
23
PDB header: transferaseChain: A: PDB Molecule: pts system, lactose-specific iibc components;PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
19 c2l2qA_
95.9
18
PDB header: transferaseChain: A: PDB Molecule: pts system, cellobiose-specific iib component (cela);PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
20 c2kyrA_
92.5
17
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
21 d1ycga1
not modelled
90.3
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related22 c3b9yA_
not modelled
88.9
14
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
23 d1hynp_
not modelled
88.7
13
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain24 d2arka1
not modelled
88.7
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like25 d2fcra_
not modelled
88.4
20
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related26 d1vmea1
not modelled
87.8
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related27 d1f4pa_
not modelled
87.8
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related28 c1hynQ_
not modelled
87.0
16
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
29 c3edoA_
not modelled
87.0
23
PDB header: flavoproteinChain: A: PDB Molecule: putative trp repressor binding protein;PDBTitle: crystal structure of flavoprotein in complex with fmn2 (yp_193882.1) from lactobacillus acidophilus ncfm at 1.203 a resolution
30 c2wc1A_
not modelled
86.6
17
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
31 c1bvyF_
not modelled
86.5
16
PDB header: oxidoreductaseChain: F: PDB Molecule: protein (cytochrome p450 bm-3);PDBTitle: complex of the heme and fmn-binding domains of the2 cytochrome p450(bm-3)
32 d1bvyf_
not modelled
86.5
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related33 c2hnbA_
not modelled
86.2
14
PDB header: electron transportChain: A: PDB Molecule: protein mioc;PDBTitle: solution structure of a bacterial holo-flavodoxin
34 d1e5da1
not modelled
86.1
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related35 d2r48a1
not modelled
86.0
16
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like36 c3f6sI_
not modelled
84.9
19
PDB header: electron transportChain: I: PDB Molecule: flavodoxin;PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
37 d1czna_
not modelled
84.8
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related38 d1yoba1
not modelled
84.8
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related39 d1tlla2
not modelled
84.8
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like40 c3rofA_
not modelled
84.4
17
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-phosphatase ptpa;PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
41 c3hr4C_
not modelled
84.2
12
PDB header: oxidoreductase/metal binding proteinChain: C: PDB Molecule: nitric oxide synthase, inducible;PDBTitle: human inos reductase and calmodulin complex
42 d1b1ca_
not modelled
82.8
19
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like43 d1y1la_
not modelled
82.8
27
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases44 c3hlyA_
not modelled
82.7
4
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
45 c2gi4A_
not modelled
81.9
16
PDB header: hydrolaseChain: A: PDB Molecule: possible phosphotyrosine protein phosphatase;PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
46 d1dg9a_
not modelled
81.7
23
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases47 d1oboa_
not modelled
80.8
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related48 d5nula_
not modelled
78.9
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related49 d5pnta_
not modelled
78.7
23
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases50 c3klbA_
not modelled
76.4
22
PDB header: flavoproteinChain: A: PDB Molecule: putative flavoprotein;PDBTitle: crystal structure of putative flavoprotein in complex with fmn2 (yp_213683.1) from bacteroides fragilis nctc 9343 at 1.75 a3 resolution
51 c3fniA_
not modelled
76.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
52 c2l18A_
not modelled
76.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: an arsenate reductase in the phosphate binding state
53 c3d7nA_
not modelled
75.1
12
PDB header: electron transportChain: A: PDB Molecule: flavodoxin, wrba-like protein;PDBTitle: the crystal structure of the flavodoxin, wrba-like protein from2 agrobacterium tumefaciens
54 c2cwdA_
not modelled
73.7
18
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight phosphotyrosine protein phosphatase;PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
55 c2ohiB_
not modelled
73.2
11
PDB header: oxidoreductaseChain: B: PDB Molecule: type a flavoprotein fpra;PDBTitle: crystal structure of coenzyme f420h2 oxidase (fpra), a diiron2 flavoprotein, reduced state
56 c3nhzA_
not modelled
72.9
23
PDB header: dna binding proteinChain: A: PDB Molecule: two component system transcriptional regulator mtra;PDBTitle: structure of n-terminal domain of mtra
57 d1fuea_
not modelled
72.1
21
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related58 c1vmeB_
not modelled
71.5
18
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
59 c1u2pA_
not modelled
71.5
13
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
60 d1ydga_
not modelled
70.1
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like61 d1jf8a_
not modelled
69.6
14
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases62 d1kgda_
not modelled
69.2
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases63 d2oara1
not modelled
68.9
9
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel64 c1wv9B_
not modelled
68.4
26
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: rhodanese homolog tt1651;PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
65 c2oarA_
not modelled
68.1
9
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
66 d2vo1a1
not modelled
67.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like67 d2fz5a1
not modelled
67.0
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related68 c2f00A_
not modelled
66.2
17
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
69 d1p3da1
not modelled
65.8
15
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain70 d2r4qa1
not modelled
64.1
15
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like71 d1ykga1
not modelled
63.6
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like72 c2wmyH_
not modelled
63.4
23
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
73 c3onoA_
not modelled
62.6
24
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
74 d1ja1a2
not modelled
61.9
24
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like75 c2ppwA_
not modelled
61.3
24
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
76 c2fekA_
not modelled
60.1
16
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
77 c3he8A_
not modelled
58.6
46
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
78 c1zggA_
not modelled
58.4
19
PDB header: hydrolaseChain: A: PDB Molecule: putative low molecular weight protein-tyrosine-PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
79 c3tovB_
not modelled
58.3
19
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase family 9;PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
80 c3m1pA_
not modelled
57.6
31
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
81 c3k7pA_
not modelled
57.6
31
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
82 d1p8aa_
not modelled
57.4
17
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases83 d2vvpa1
not modelled
57.4
24
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB84 d1d1qa_
not modelled
57.3
12
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases85 d1nn4a_
not modelled
57.2
38
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB86 c3s5pA_
not modelled
56.7
31
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
87 c3qd5B_
not modelled
55.1
22
PDB header: isomeraseChain: B: PDB Molecule: putative ribose-5-phosphate isomerase;PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
88 c3jviA_
not modelled
53.7
19
PDB header: hydrolaseChain: A: PDB Molecule: protein tyrosine phosphatase;PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
89 d1x9ia_
not modelled
52.9
20
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain90 c3cz5B_
not modelled
52.6
14
PDB header: transcription regulatorChain: B: PDB Molecule: two-component response regulator, luxr family;PDBTitle: crystal structure of two-component response regulator, luxr family,2 from aurantimonas sp. si85-9a1
91 c2jrlA_
not modelled
50.3
9
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
92 d1j6ua1
not modelled
48.7
17
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain93 c3b6iB_
not modelled
48.4
15
PDB header: flavoproteinChain: B: PDB Molecule: flavoprotein wrba;PDBTitle: wrba from escherichia coli, native structure
94 d1n1ea2
not modelled
47.7
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain95 c3snkA_
not modelled
47.4
12
PDB header: signaling proteinChain: A: PDB Molecule: response regulator chey-like protein;PDBTitle: crystal structure of a response regulator chey-like protein (mll6475)2 from mesorhizobium loti at 2.02 a resolution
96 d2eyqa5
not modelled
46.9
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain97 d1g16a_
not modelled
46.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins98 c3c5yD_
not modelled
45.5
36
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
99 d1s8na_
not modelled
43.8
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related100 d1yt8a4
not modelled
43.6
17
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)101 c3tp9B_
not modelled
42.7
34
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase and rhodanese domain protein;PDBTitle: crystal structure of alicyclobacillus acidocaldarius protein with2 beta-lactamase and rhodanese domains
102 d1o1xa_
not modelled
41.1
27
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB103 c3ilmD_
not modelled
39.9
22
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr3790 protein;PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
104 c3fojA_
not modelled
39.9
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ssp1007 from staphylococcus2 saprophyticus subsp. saprophyticus. northeast structural3 genomics target syr101a.
105 c3jteA_
not modelled
39.7
14
PDB header: protein bindingChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver domain2 protein from clostridium thermocellum
106 c3t38B_
not modelled
39.1
16
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
107 d1jl3a_
not modelled
39.1
22
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases108 c3mb8A_
not modelled
39.0
17
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of purine nucleoside phosphorylase from toxoplasma2 gondii in complex with immucillin-h
109 d1q1aa_
not modelled
38.6
15
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators110 c3bjeA_
not modelled
37.5
23
PDB header: transferaseChain: A: PDB Molecule: nucleoside phosphorylase, putative;PDBTitle: crystal structure of trypanosoma brucei nucleoside phosphorylase shows2 uridine phosphorylase activity
111 c1ir6A_
not modelled
35.6
16
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease recj;PDBTitle: crystal structure of exonuclease recj bound to manganese
112 d1ir6a_
not modelled
35.6
16
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Exonuclease RecJ113 d1v3aa_
not modelled
35.2
26
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like114 c3r2uC_
not modelled
34.8
14
PDB header: hydrolaseChain: C: PDB Molecule: metallo-beta-lactamase family protein;PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
115 c3tl6B_
not modelled
34.7
22
PDB header: transferaseChain: B: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of purine nucleoside phosphorylase from entamoeba2 histolytica
116 c3i2vA_
not modelled
34.1
22
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase and sulfurtransferase mocs3;PDBTitle: crystal structure of human mocs3 rhodanese-like domain
117 d1ag9a_
not modelled
33.9
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related118 d1je0a_
not modelled
33.6
10
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases119 c1gqqA_
not modelled
32.9
14
PDB header: cell wall biosynthesisChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
120 d1vi2a1
not modelled
32.4
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain