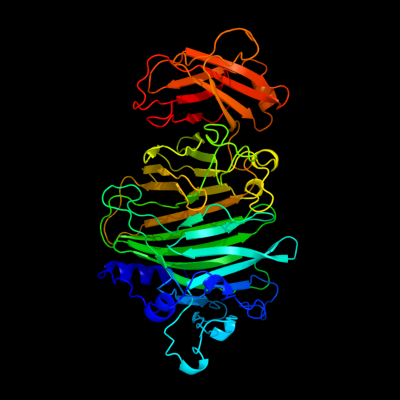
1 c1txkA_
100.0
36
PDB header: biosynthetic proteinChain: A: PDB Molecule: glucans biosynthesis protein g;PDBTitle: crystal structure of escherichia coli opgg
2 d1txka2
100.0
41
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: MdoG-like3 d1txka1
100.0
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes4 c2e76D_
53.0
11
PDB header: photosynthesisChain: D: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
5 c2r5oA_
44.6
14
PDB header: transport proteinChain: A: PDB Molecule: putative atp binding component of abc-PDBTitle: crystal structure of the c-terminal domain of wzt
6 d1nkra1
21.4
44
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains7 d2d7na1
20.6
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)8 d2jnaa1
20.4
17
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like9 d2noca1
19.6
14
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like10 c1p84E_
18.5
9
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: hdbt inhibited yeast cytochrome bc1 complex
11 c1se9A_
17.8
14
PDB header: plant proteinChain: A: PDB Molecule: ubiquitin family;PDBTitle: structure of at3g01050, a ubiquitin-fold protein from2 arabidopsis thaliana
12 d1se9a_
17.8
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related13 c3on7C_
17.4
17
PDB header: oxidoreductaseChain: C: PDB Molecule: oxidoreductase, iron/ascorbate family;PDBTitle: crystal structure of a putative oxygenase (so_2589) from shewanella2 oneidensis at 2.20 a resolution
14 c3rhfB_
17.2
13
PDB header: transferaseChain: B: PDB Molecule: putative polyphosphate kinase 2 family protein;PDBTitle: crystal structure of polyphosphate kinase 2 from arthrobacter2 aurescens tc1
15 c2l6mA_
16.5
38
PDB header: hydrolaseChain: A: PDB Molecule: protein dicer;PDBTitle: structure of c-terminal dsrbd of the fission yeast dicer (dcr1)
16 d1oisa_
15.6
24
Fold: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragmentSuperfamily: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragmentFamily: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment17 d1jmxa5
15.3
13
Fold: Streptavidin-likeSuperfamily: Quinohemoprotein amine dehydrogenase A chain, domain 3Family: Quinohemoprotein amine dehydrogenase A chain, domain 318 c2r3aA_
15.0
17
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv39h2;PDBTitle: methyltransferase domain of human suppressor of variegation2 3-9 homolog 2
19 d2dl2a1
15.0
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains20 d1xbwa_
13.7
32
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: PG130-like21 c3r0eC_
not modelled
12.8
16
PDB header: sugar binding proteinChain: C: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
22 c3ooxA_
not modelled
12.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: putative 2og-fe(ii) oxygenase family protein;PDBTitle: crystal structure of a putative 2og-fe(ii) oxygenase family protein2 (cc_0200) from caulobacter crescentus at 1.44 a resolution
23 d1dmla1
not modelled
11.9
26
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor24 c3exmA_
not modelled
11.7
24
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase sc4828;PDBTitle: crystal structure of the phosphatase sc4828 with the non-hydrolyzable2 nucleotide gpcp
25 c2l3bA_
not modelled
11.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
26 d1k4ta3
not modelled
11.4
24
Fold: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragmentSuperfamily: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragmentFamily: Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment27 c2i1jA_
not modelled
11.4
35
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda at 2.1 angstroms resolution
28 c2fynO_
not modelled
11.3
17
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
29 c2l7qA_
not modelled
11.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
30 d1ee8a2
not modelled
11.3
22
Fold: N-terminal domain of MutM-like DNA repair proteinsSuperfamily: N-terminal domain of MutM-like DNA repair proteinsFamily: N-terminal domain of MutM-like DNA repair proteins31 d1csya_
not modelled
11.0
18
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain32 c2fyuE_
not modelled
10.8
22
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit,PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
33 d1qcsa1
not modelled
10.2
43
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like34 c3phuA_
not modelled
10.2
9
PDB header: hydrolaseChain: A: PDB Molecule: rna-directed rna polymerase l;PDBTitle: otu domain of crimean congo hemorrhagic fever virus
35 d1ix2a_
not modelled
9.8
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)36 d1ml9a_
not modelled
9.4
13
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases37 c2p8zS_
not modelled
9.3
56
PDB header: translationChain: S: PDB Molecule: elongation factor tu-b;PDBTitle: fitted structure of adpr-eef2 in the 80s:adpr-2 eef2:gdpnp:sordarin cryo-em reconstruction
38 d1erna1
not modelled
9.2
34
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III39 d1h95a_
not modelled
9.2
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like40 d1e09a_
not modelled
9.1
23
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like41 d1p1la_
not modelled
9.0
40
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)42 d1ja1a3
not modelled
9.0
44
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like43 c3d3zA_
not modelled
8.9
27
PDB header: hydrolaseChain: A: PDB Molecule: actibind;PDBTitle: crystal structure of actibind a t2 rnase
44 c3o0lB_
not modelled
8.7
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
45 c3camB_
not modelled
8.7
56
PDB header: gene regulationChain: B: PDB Molecule: cold-shock domain family protein;PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
46 d1a8pa2
not modelled
8.6
44
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases47 d1fdra2
not modelled
8.5
33
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases48 c1a31A_
not modelled
8.4
24
PDB header: isomerase/dnaChain: A: PDB Molecule: protein (topoisomerase i);PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
49 c1dmlG_
not modelled
8.4
26
PDB header: dna binding protein/transferaseChain: G: PDB Molecule: dna polymerase processivity factor;PDBTitle: crystal structure of herpes simplex ul42 bound to the c-2 terminus of hsv pol
50 d1dq3a3
not modelled
8.2
40
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease51 c3adyA_
not modelled
8.2
29
PDB header: proton transportChain: A: PDB Molecule: dotd;PDBTitle: crystal structure of dotd from legionella
52 d1ef1a2
not modelled
8.2
35
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM53 c3nezA_
not modelled
8.1
22
PDB header: fluorescent proteinChain: A: PDB Molecule: mrojoa;PDBTitle: mrojoa
54 d1kr4a_
not modelled
7.9
10
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)55 d1b2pa_
not modelled
7.9
19
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins56 c1ckwA_
not modelled
7.8
12
PDB header: metal transportChain: A: PDB Molecule: protein (cystic fibrosis transmembranePDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
57 c1nh3A_
not modelled
7.8
24
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase i;PDBTitle: human topoisomerase i ara-c complex
58 c2rocB_
not modelled
7.8
33
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-binding component 3;PDBTitle: solution structure of mcl-1 complexed with puma
59 d1f20a2
not modelled
7.8
56
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like60 d2ozga1
not modelled
7.7
26
Fold: SCP-likeSuperfamily: SCP-likeFamily: EF1021 C-terminal domain-like61 d1oq1a_
not modelled
7.7
16
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Hypothetical protein YesU62 d1ukua_
not modelled
7.6
0
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)63 d1pjca1
not modelled
7.6
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain64 d2oq1a2
not modelled
7.5
9
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain65 c2k7pA_
not modelled
7.5
18
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: filamin a ig-like domains 16-17
66 d2a5yb1
not modelled
7.5
47
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CED-4 C-terminal domain-like67 c2qsdB_
not modelled
7.5
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of a protein il1583 from idiomarina loihiensis
68 d1kbia1
not modelled
7.4
16
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases69 c3czpA_
not modelled
7.4
21
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from pseudomonas2 aeruginosa pa01
70 c2ns2A_
not modelled
7.3
29
PDB header: cell cycleChain: A: PDB Molecule: spindlin-1;PDBTitle: crystal structure of spindlin1
71 d1ddga2
not modelled
7.3
44
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like72 c2wclA_
not modelled
7.3
36
PDB header: transport proteinChain: A: PDB Molecule: general odorant-binding protein 1;PDBTitle: structure of bmori gobp2 (general odorant binding protein 2)2 with (8e,10z)-hexadecadien-1-ol
73 d1p7hl1
not modelled
7.3
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain74 c2kl5A_
not modelled
7.3
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yutd;PDBTitle: solution nmr structure of protein yutd from b.subtilis, northeast2 structural genomics consortium target sr232
75 c3a0jB_
not modelled
7.2
33
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
76 c2b9sA_
not modelled
7.2
29
PDB header: isomerase/dnaChain: A: PDB Molecule: topoisomerase i-like protein;PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
77 d1d6za3
not modelled
7.2
17
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region78 d1imhc1
not modelled
7.1
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain79 d1dlpa2
not modelled
7.1
24
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins80 d1nkra2
not modelled
7.0
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains81 c1wrgA_
not modelled
6.8
80
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, beta chain;PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
82 c1ibtC_
not modelled
6.8
23
PDB header: lyaseChain: C: PDB Molecule: histidine decarboxylase beta chain;PDBTitle: structure of the d53,54n mutant of histidine decarboxylase at-170 c
83 c3r2nC_
not modelled
6.7
16
PDB header: hydrolaseChain: C: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium leprae
84 c2jp2A_
not modelled
6.7
13
PDB header: signaling proteinChain: A: PDB Molecule: sprouty-related, evh1 domain-containing proteinPDBTitle: solution structure and resonance assignment of the n-2 terminal evh1 domain from the human spred2 protein3 (sprouty-related protein with evh1 domain isoform 2)
85 d2bk0a1
not modelled
6.7
27
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like86 d1kfia4
not modelled
6.6
21
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain87 d1my7a_
not modelled
6.6
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain88 c1meqA_
not modelled
6.6
56
PDB header: viral proteinChain: A: PDB Molecule: exterior membrane glycoprotein (gp120);PDBTitle: hiv gp120 c5
89 d2k54a1
not modelled
6.6
14
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Atu0742-like90 c2eiuE_
not modelled
6.6
31
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: hypothetical protein aq_1627;PDBTitle: crystal structure of a putative protein (aq1627) from aquifex aeolicus
91 c1ckxA_
not modelled
6.6
10
PDB header: metal transportChain: A: PDB Molecule: cystic fibrosis transmembrane conductancePDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
92 d1h4ra2
not modelled
6.6
35
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM93 c2kvhA_
not modelled
6.5
63
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
94 c2a48A_
not modelled
6.5
20
PDB header: luminescent proteinChain: A: PDB Molecule: gfp-like fluorescent chromoprotein amfp486;PDBTitle: crystal structure of amfp486 e150q
95 d1m4ka2
not modelled
6.5
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains96 c3pt2A_
not modelled
6.5
11
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: rna polymerase;PDBTitle: structure of a viral otu domain protease bound to ubiquitin
97 d1x9na2
not modelled
6.4
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain98 d1gawa2
not modelled
6.4
56
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases99 d1jb9a2
not modelled
6.4
33
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases