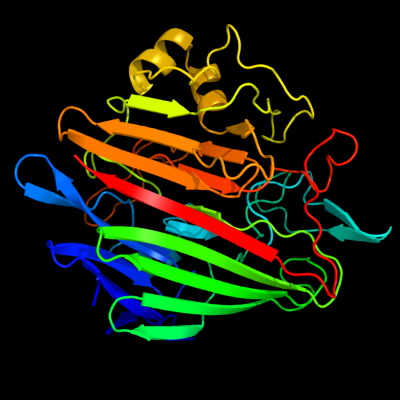
1 d1so0a_
100.0
37
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)2 c1z45A_
100.0
25
PDB header: isomeraseChain: A: PDB Molecule: gal10 bifunctional protein;PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
3 d1lura_
100.0
33
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)4 d1nsza_
100.0
31
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)5 c3imhB_
100.0
34
PDB header: isomeraseChain: B: PDB Molecule: galactose-1-epimerase;PDBTitle: crystal structure of galactose 1-epimerase from lactobacillus2 acidophilus ncfm
6 d1z45a1
100.0
25
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)7 c1ygaA_
100.0
26
PDB header: isomeraseChain: A: PDB Molecule: hypothetical 37.9 kda protein in bio3-hxt17PDBTitle: crystal structure of saccharomyces cerevisiae yn9a protein,2 new york structural genomics consortium
8 c3os7D_
100.0
14
PDB header: isomeraseChain: D: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
9 c3os7B_
100.0
14
PDB header: isomeraseChain: B: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
10 c3nreB_
100.0
19
PDB header: isomeraseChain: B: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative aldose 1-epimerase (b2544) from2 escherichia coli k12 at 1.59 a resolution
11 c3dcdA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: x-ray structure of the galactose mutarotase related enzyme q5fkd7 from2 lactobacillus acidophilus at the resolution 1.9a. northeast3 structural genomics consortium target lar33.
12 c3q1nA_
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: crystal structure of a galactose mutarotase-like protein (lsei_2598)2 from lactobacillus casei atcc 334 at 1.61 a resolution
13 c3mwxA_
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution
14 c3k25B_
100.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1438 protein;PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
15 c2cisA_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: hexose-6-phosphate mutarotase;PDBTitle: structure-based functional annotation: yeast ymr099c codes2 for a d-hexose-6-phosphate mutarotase. complex with3 tagatose-6-phosphate
16 c2htbB_
100.0
16
PDB header: isomeraseChain: B: PDB Molecule: putative enzyme related to aldose 1-epimerase;PDBTitle: crystal structure of a putative mutarotase (yead) from2 salmonella typhimurium in monoclinic form
17 d1jova_
100.0
18
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hypothetical protein HI131718 c3ty1B_
99.9
13
PDB header: isomeraseChain: B: PDB Molecule: hypothetical aldose 1-epimerase;PDBTitle: crystal structure of a hypothetical aldose 1-epimerase (kpn_04629)2 from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.90 a3 resolution
19 c3bs6B_
97.3
17
PDB header: membrane protein, protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: 1.8 angstrom crystal structure of the periplasmic domain of2 the membrane insertase yidc
20 c3blcB_
96.2
17
PDB header: chaperone,protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: crystal structure of the periplasmic domain of the escherichia coli2 yidc
21 c1so9A_
not modelled
59.9
24
PDB header: metal transportChain: A: PDB Molecule: cytochrome c oxidase assembly protein ctag;PDBTitle: solution structure of apocox11, 30 structures
22 d1so9a_
not modelled
59.9
24
Fold: Ctag/Cox11Superfamily: Ctag/Cox11Family: Ctag/Cox1123 d1k1xa2
not modelled
45.1
13
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: 4-alpha-glucanotransferase, C-terminal domain24 d1jz8a4
not modelled
19.0
20
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 525 c2xn1B_
not modelled
18.6
12
PDB header: hydrolaseChain: B: PDB Molecule: alpha-galactosidase;PDBTitle: structure of alpha-galactosidase from lactobacillus acidophilus ncfm2 with tris
26 d2hkja2
not modelled
14.6
45
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain27 c2zooA_
not modelled
14.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nitrite reductase;PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
28 d1f2ri_
not modelled
13.9
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain29 d4ubpb_
not modelled
12.8
18
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit30 d1oe1a1
not modelled
12.8
14
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins31 d1e9ya1
not modelled
12.2
18
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit32 d1ibxb_
not modelled
10.0
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain33 c1ibxB_
not modelled
10.0
25
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: chimera of igg binding protein g and dnaPDBTitle: nmr structure of dff40 and dff45 n-terminal domain complex
34 d1c9fa_
not modelled
9.9
32
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain35 d1q2la4
not modelled
9.9
29
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like36 d1ejxb_
not modelled
9.5
17
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit37 d1ibxa_
not modelled
9.3
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain38 c2zf9D_
not modelled
8.9
6
PDB header: structural proteinChain: D: PDB Molecule: scae cell-surface anchored scaffoldin protein;PDBTitle: crystal structure of a type iii cohesin module from the cellulosomal2 scae cell-surface anchoring scaffoldin of ruminococcus flavefaciens
39 d1ndsa1
not modelled
8.1
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins40 c3cw3A_
not modelled
8.0
16
PDB header: oncoproteinChain: A: PDB Molecule: absent in melanoma 1 protein;PDBTitle: crystal structure of aim1g1
41 c2g8gA_
not modelled
7.2
40
PDB header: virusChain: A: PDB Molecule: capsid;PDBTitle: structurally mapping the diverse phenotype of adeno-2 associated virus serotype 4
42 d1m53a1
not modelled
7.0
9
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain43 c3mi6A_
not modelled
6.5
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of the alpha-galactosidase from lactobacillus2 brevis, northeast structural genomics consortium target lbr11.
44 c3kieF_
not modelled
6.4
33
PDB header: virusChain: F: PDB Molecule: capsid protein vp1;PDBTitle: crystal structure of adeno-associated virus serotype 3b
45 c1v06A_
not modelled
6.3
33
PDB header: dna-binding proteinChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: axh domain of the transcription factor hbp1 from m.musculus
46 d1yq2a5
not modelled
6.2
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases47 c2x3bB_
not modelled
6.2
34
PDB header: hydrolaseChain: B: PDB Molecule: toxic extracellular endopeptidase;PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
48 c2kl8A_
not modelled
6.1
44
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
49 c1wa1X_
not modelled
6.0
14
PDB header: reductaseChain: X: PDB Molecule: dissimilatory copper-containing nitritePDBTitle: crystal structure of h313q mutant of alcaligenes2 xylosoxidans nitrite reductase
50 c3nttA_
not modelled
5.9
39
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: structural insights of adeno-associated virus 5. a gene therapy vector2 for cystic fibrosis
51 c3dqgC_
not modelled
5.9
11
PDB header: chaperoneChain: C: PDB Molecule: heat shock 70 kda protein f;PDBTitle: peptide-binding domain of heat shock 70 kda protein f, mitochondrial2 precursor, from caenorhabditis elegans.
52 d1s58a_
not modelled
5.5
29
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: ssDNA virusesFamily: Parvoviridae-like VP53 c2eelA_
not modelled
5.4
22
PDB header: apoptosisChain: A: PDB Molecule: cell death activator cide-a;PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
54 c3qgaD_
not modelled
5.4
17
PDB header: hydrolaseChain: D: PDB Molecule: fusion of urease beta and gamma subunits;PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
55 d1mzya1
not modelled
5.3
14
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins56 d1vlba2
not modelled
5.3
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins