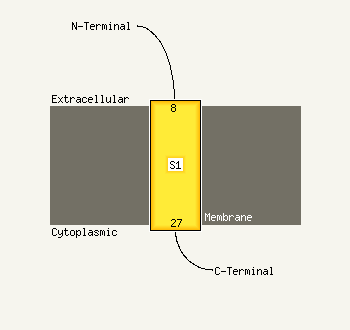
1 d1hnga1
46.1
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)2 c2kr6A_
45.3
31
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
3 d1v54c_
36.6
27
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like4 c1q8hA_
32.5
45
PDB header: metal binding proteinChain: A: PDB Molecule: osteocalcin;PDBTitle: crystal structure of porcine osteocalcin
5 d1q8ha_
32.5
45
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain6 d1ikpa1
30.5
18
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Exotoxin A, N-terminal domain7 c3k3gA_
29.9
21
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
8 c3kp9A_
26.5
10
PDB header: blood coagulation,oxidoreductaseChain: A: PDB Molecule: vkorc1/thioredoxin domain protein;PDBTitle: structure of a bacterial homolog of vitamin k epoxide reductase
9 d1hnfa1
26.3
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)10 d1q3ma_
25.3
45
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain11 c2vesA_
21.5
17
PDB header: hydrolaseChain: A: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: crystal structure of lpxc from pseudomonas aeruginosa2 complexed with the potent bb-78485 inhibitor
12 d1oria_
21.2
12
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Arginine methyltransferase13 c3nzkB_
20.3
17
PDB header: hydrolaseChain: B: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: structure of lpxc from yersinia enterocolitica complexed with chir0902 inhibitor
14 d1qlec_
20.0
23
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like15 c2yboA_
19.9
21
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
16 d1e2wa1
19.4
12
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain17 d1g47a1
18.3
40
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain18 d1yvra1
17.0
33
Fold: alpha-alpha superhelixSuperfamily: TROVE domain-likeFamily: TROVE domain-like19 d1vf5c1
16.9
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain20 d1k0ha_
16.7
9
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: gpFII-like21 c3o44G_
not modelled
15.9
18
PDB header: toxinChain: G: PDB Molecule: hemolysin;PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya) heptameric2 pore
22 c1xezA_
not modelled
15.6
18
PDB header: toxinChain: A: PDB Molecule: hemolysin;PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya)2 pro-toxin with octylglucoside bound
23 c1e2vB_
not modelled
15.3
12
PDB header: electron transport proteinsChain: B: PDB Molecule: cytochrome f;PDBTitle: n153q mutant of cytochrome f from chlamydomonas reinhardtii
24 c3c5oD_
not modelled
15.2
9
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: upf0311 protein rpa1785;PDBTitle: crystal structure of the conserved protein of unknown function rpa17852 from rhodopseudomonas palustris
25 c2v1lA_
not modelled
14.8
23
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of the conserved hypothetical protein vc1805 from2 pathogenicity island vpi-2 of vibrio cholerae o1 biovar3 eltor str. n16961 shares structural homology with the4 human p32 protein
26 c2p8zS_
not modelled
14.7
63
PDB header: translationChain: S: PDB Molecule: elongation factor tu-b;PDBTitle: fitted structure of adpr-eef2 in the 80s:adpr-2 eef2:gdpnp:sordarin cryo-em reconstruction
27 c1q90A_
not modelled
14.4
12
PDB header: photosynthesisChain: A: PDB Molecule: apocytochrome f;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
28 c2rddB_
not modelled
13.7
6
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
29 c1ikqA_
not modelled
13.6
18
PDB header: transferaseChain: A: PDB Molecule: exotoxin a;PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
30 d1ci3m1
not modelled
13.6
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain31 c3g7gG_
not modelled
13.4
17
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: upf0311 protein ca_c3321;PDBTitle: crystal structure of the protein with unknown function from2 clostridium acetobutylicum atcc 824
32 d1tu2b1
not modelled
12.6
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain33 c2w56B_
not modelled
12.6
20
PDB header: unknown functionChain: B: PDB Molecule: vc0508;PDBTitle: structure of the hypothetical protein vc0508 from vibrio cholerae2 vsp-ii pathogenicity island
34 d1hcza1
not modelled
11.9
12
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain35 c3dwuA_
not modelled
11.8
63
PDB header: biosynthetic proteinChain: A: PDB Molecule: elongation factor tu-b;PDBTitle: transition-state model conformation of the switch i region2 fitted into the cryo-em map of the eef2.80s.alf4.gdp3 complex
36 c3demB_
not modelled
11.6
6
PDB header: hydrolaseChain: B: PDB Molecule: complement factor masp-3;PDBTitle: cub1-egf-cub2 domain of human masp-1/3
37 c2e75C_
not modelled
11.6
15
PDB header: photosynthesisChain: C: PDB Molecule: apocytochrome f;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
38 d1iwga8
not modelled
11.5
13
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain39 c2k9pA_
not modelled
11.5
25
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
40 d3c8ya2
not modelled
11.1
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins41 d1v97a1
not modelled
10.5
46
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like42 d1jb0l_
not modelled
10.5
27
Fold: Photosystem I reaction center subunit XI, PsaLSuperfamily: Photosystem I reaction center subunit XI, PsaLFamily: Photosystem I reaction center subunit XI, PsaL43 c1ctmA_
not modelled
10.5
14
PDB header: electron transport(cytochrome)Chain: A: PDB Molecule: cytochrome f;PDBTitle: crystal structure of chloroplast cytochrome f reveals a2 novel cytochrome fold and unexpected heme ligation
44 d1mtyg_
not modelled
10.3
27
Fold: Open three-helical up-and-down bundleSuperfamily: Methane monooxygenase hydrolase, gamma subunitFamily: Methane monooxygenase hydrolase, gamma subunit45 d3cx5e2
not modelled
9.8
24
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor46 c2k6zA_
not modelled
9.8
25
PDB header: metal transportChain: A: PDB Molecule: putative uncharacterized protein ttha1943;PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
47 d1m56c_
not modelled
9.6
15
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like48 d1mhyg_
not modelled
9.5
33
Fold: Open three-helical up-and-down bundleSuperfamily: Methane monooxygenase hydrolase, gamma subunitFamily: Methane monooxygenase hydrolase, gamma subunit49 c2nvoA_
not modelled
9.4
33
PDB header: rna binding proteinChain: A: PDB Molecule: ro sixty-related protein, rsr;PDBTitle: crystal structure of deinococcus radiodurans ro (rsr) protein
50 c2gutA_
not modelled
9.1
25
PDB header: transcriptionChain: A: PDB Molecule: arc/mediator, positive cofactor 2 glutamine/q-PDBTitle: solution structure of the trans-activation domain of the2 human co-activator arc105
51 c3lrcC_
not modelled
8.9
11
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
52 c1dnuB_
not modelled
8.8
23
PDB header: oxidoreductaseChain: B: PDB Molecule: myeloperoxidase;PDBTitle: structural analyses of human myeloperoxidase-thiocyanate complex
53 c2oarA_
not modelled
8.7
17
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
54 d1k92a2
not modelled
8.6
30
Fold: Argininosuccinate synthetase, C-terminal domainSuperfamily: Argininosuccinate synthetase, C-terminal domainFamily: Argininosuccinate synthetase, C-terminal domain55 c2iv1J_
not modelled
8.4
36
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
56 c1orhA_
not modelled
8.4
13
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 1;PDBTitle: structure of the predominant protein arginine methyltransferase prmt1
57 c2jxmB_
not modelled
8.2
12
PDB header: electron transportChain: B: PDB Molecule: cytochrome f;PDBTitle: ensemble of twenty structures of the prochlorothrix2 hollandica plastocyanin- cytochrome f complex
58 d1x9la_
not modelled
8.1
25
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: DR1885-like metal-binding proteinFamily: DR1885-like metal-binding protein59 c1tu2B_
not modelled
7.8
15
PDB header: electron transportChain: B: PDB Molecule: apocytochrome f;PDBTitle: the complex of nostoc cytochrome f and plastocyanin determin with2 paramagnetic nmr. based on the structures of cytochrome f and3 plastocyanin, 10 structures
60 d1vava_
not modelled
7.7
25
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Alginate lyase61 d2i5nl1
not modelled
7.7
14
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits62 c3c25A_
not modelled
7.6
21
PDB header: hydrolase/dnaChain: A: PDB Molecule: noti restriction endonuclease;PDBTitle: crystal structure of noti restriction endonuclease bound to cognate2 dna
63 c2l4wA_
not modelled
7.4
29
PDB header: protein transportChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the xanthomonas virb7
64 d1k8ba_
not modelled
7.4
18
Fold: Ribosome binding domain-likeSuperfamily: Translation initiation factor 2 beta, aIF2beta, N-terminal domainFamily: Translation initiation factor 2 beta, aIF2beta, N-terminal domain65 d2f41a1
not modelled
7.3
15
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: PaaI/YdiI-like66 d1t3qa1
not modelled
7.1
20
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like67 c2kqtD_
not modelled
7.1
26
PDB header: transport proteinChain: D: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
68 c2kqtA_
not modelled
7.1
26
PDB header: transport proteinChain: A: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
69 c2kqtB_
not modelled
7.1
26
PDB header: transport proteinChain: B: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
70 c2kqtC_
not modelled
7.1
26
PDB header: transport proteinChain: C: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
71 d1mv8a1
not modelled
7.1
36
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: UDP-glucose/GDP-mannose dehydrogenase dimerisation domain72 d1nt0a2
not modelled
7.1
13
Fold: CUB-likeSuperfamily: Spermadhesin, CUB domainFamily: Spermadhesin, CUB domain73 c2lhuA_
not modelled
7.0
50
PDB header: structural proteinChain: A: PDB Molecule: mybpc3 protein;PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
74 c2yvxD_
not modelled
7.0
17
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
75 d2axth1
not modelled
6.9
28
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like76 d2oiea1
not modelled
6.9
58
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: MazG-like77 c3o0rB_
not modelled
6.8
6
PDB header: immune system/oxidoreductaseChain: B: PDB Molecule: nitric oxide reductase subunit b;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
78 d1zxia1
not modelled
6.8
25
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like79 c1nyjC_
not modelled
6.8
28
PDB header: viral proteinChain: C: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
80 c1mp6A_
not modelled
6.8
28
PDB header: membrane proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: structure of the transmembrane region of the m2 protein h+2 channel by solid state nmr spectroscopy
81 c1nyjB_
not modelled
6.8
28
PDB header: viral proteinChain: B: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
82 c1nyjA_
not modelled
6.8
28
PDB header: viral proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
83 c1nyjD_
not modelled
6.8
28
PDB header: viral proteinChain: D: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
84 c3pojA_
not modelled
6.7
7
PDB header: hydrolaseChain: A: PDB Molecule: mannan-binding lectin serine protease 1;PDBTitle: crystal structure of masp-1 cub2 domain bound to ethylamine
85 d2c5ra1
not modelled
6.7
25
Fold: Phage replication organizer domainSuperfamily: Phage replication organizer domainFamily: Phage replication organizer domain86 d1jroa1
not modelled
6.6
50
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like87 c1ds5F_
not modelled
6.6
45
PDB header: transferaseChain: F: PDB Molecule: casein kinase, beta chain;PDBTitle: dimeric crystal structure of the alpha subunit in complex2 with two beta peptides mimicking the architecture of the3 tetrameric protein kinase ck2 holoenzyme.
88 c2q4pA_
not modelled
6.6
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein rs21-c6;PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898
89 d2a3qa1
not modelled
6.6
32
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: MazG-like90 c2q9lA_
not modelled
6.4
42
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of imazg from vibrio dat 722: ctag-imazg (p43212)
91 c2jttD_
not modelled
6.3
13
PDB header: calcium binding protein/antitumor proteiChain: D: PDB Molecule: calcyclin-binding protein;PDBTitle: solution structure of calcium loaded s100a6 bound to c-2 terminal siah-1 interacting protein
92 d2axti1
not modelled
6.3
23
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like93 c3nfgG_
not modelled
6.3
34
PDB header: transcriptionChain: G: PDB Molecule: dna-directed rna polymerase i subunit rpa49;PDBTitle: crystal structure of dimerization module of rna polymerase i2 subcomplex a49/a34.5
94 c3s88J_
not modelled
6.3
20
PDB header: immune system/viral proteinChain: J: PDB Molecule: envelope glycoprotein;PDBTitle: crystal structure of sudan ebolavirus glycoprotein (strain gulu) bound2 to 16f6
95 c3dh4A_
not modelled
6.3
8
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
96 c2f3xA_
not modelled
6.2
15
PDB header: gene regulationChain: A: PDB Molecule: transcription factor fapr;PDBTitle: crystal structure of fapr (in complex with effector)- a2 global regulator of fatty acid biosynthesis in b. subtilis
97 c2qmaB_
not modelled
6.1
25
PDB header: transferaseChain: B: PDB Molecule: diaminobutyrate-pyruvate transaminase and l-2,4-PDBTitle: crystal structure of glutamate decarboxylase domain of2 diaminobutyrate-pyruvate transaminase and l-2,4-diaminobutyrate3 decarboxylase from vibrio parahaemolyticus
98 c3rceA_
not modelled
6.1
14
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
99 c1yvrA_
not modelled
6.1
33
PDB header: rna binding proteinChain: A: PDB Molecule: 60-kda ss-a/ro ribonucleoprotein;PDBTitle: ro autoantigen