| 1 | c3giaA_
|
|
 |
100.0 |
18 |
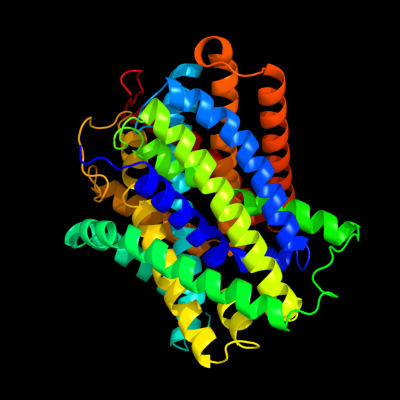
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
|
| 2 | c3lrcC_
|
|
 |
100.0 |
17 |
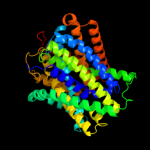
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
|
| 3 | c2jlnA_
|
|
 |
100.0 |
10 |
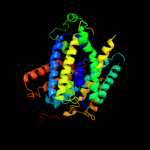
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
| 4 | c2xq2A_
|
|
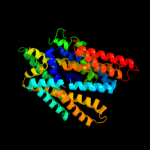 |
99.1 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
|
| 5 | c3dh4A_
|
|
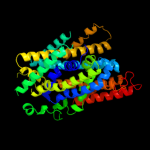 |
99.1 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
|
| 6 | c2w8aC_
|
|
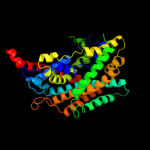 |
94.6 |
11 |
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
|
| 7 | d2a65a1
|
|
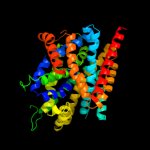 |
91.6 |
11 |
Fold:SNF-like
Superfamily:SNF-like
Family:SNF-like |
| 8 | c3hfxA_
|
|
 |
78.7 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
|
| 9 | c3qnqD_
|
|
 |
51.3 |
17 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
| 10 | d1fftb2
|
|
 |
46.2 |
15 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 11 | c2kluA_
|
|
 |
37.6 |
12 |
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
|
| 12 | c2kvtA_
|
|
 |
11.0 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yaia;
PDBTitle: solution nmr structure of yaia from escherichia eoli. northeast2 structural genomics target er244
|
| 13 | c3le4A_
|
|
 |
10.9 |
27 |
PDB header:nuclear protein
Chain: A: PDB Molecule:microprocessor complex subunit dgcr8;
PDBTitle: crystal structure of the dgcr8 dimerization domain
|
| 14 | d1qhda2
|
|
 |
10.7 |
32 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Top domain of virus capsid protein |
| 15 | c1ujlA_
|
|
 |
10.3 |
0 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
|
| 16 | c3rkoF_
|
|
 |
9.6 |
12 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
|
| 17 | c1zgxB_
|
|
 |
9.2 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:guanyl-specific ribonuclease sa;
PDBTitle: crystal structure of ribonuclease mutant
|
| 18 | c3bcyA_
|
|
 |
8.8 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:protein yer067w;
PDBTitle: crystal structure of yer067w
|
| 19 | c2l1nA_
|
|
 |
8.5 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the protein yp_399305.1
|
| 20 | c2elmA_
|
|
 |
8.4 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 10th c2h2 zinc finger of human2 zinc finger protein 406
|
| 21 | c2rddB_ |
|
not modelled |
8.0 |
22 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
|
| 22 | c2v6xB_ |
|
not modelled |
7.8 |
25 |
PDB header:protein transport
Chain: B: PDB Molecule:doa4-independent degradation protein 4;
PDBTitle: stractural insight into the interaction between escrt-iii2 and vps4
|
| 23 | d1eg3a3 |
|
not modelled |
7.6 |
33 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 24 | c3m7bA_ |
|
not modelled |
7.4 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
|
| 25 | c2owrD_ |
|
not modelled |
7.4 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:uracil-dna glycosylase;
PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
|
| 26 | c2wmpB_ |
|
not modelled |
7.3 |
29 |
PDB header:chaperone
Chain: B: PDB Molecule:papg protein;
PDBTitle: structure of the e. coli chaperone papd in complex with the pilin2 domain of the papgii adhesin
|
| 27 | d1p35a_ |
|
not modelled |
7.3 |
23 |
Fold:Baculovirus p35 protein
Superfamily:Baculovirus p35 protein
Family:Baculovirus p35 protein |
| 28 | c2i6kA_ |
|
not modelled |
7.2 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:isopentenyl-diphosphate delta-isomerase 1;
PDBTitle: crystal structure of human type i ipp isomerase complexed2 with a substrate analog
|
| 29 | d2r6gf1 |
|
not modelled |
7.2 |
14 |
Fold:MalF N-terminal region-like
Superfamily:MalF N-terminal region-like
Family:MalF N-terminal region-like |
| 30 | d2oara1 |
|
not modelled |
6.8 |
10 |
Fold:Gated mechanosensitive channel
Superfamily:Gated mechanosensitive channel
Family:Gated mechanosensitive channel |
| 31 | c2gfpA_ |
|
not modelled |
6.8 |
6 |
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
|
| 32 | d2iuba2 |
|
not modelled |
6.7 |
5 |
Fold:Transmembrane helix hairpin
Superfamily:Magnesium transport protein CorA, transmembrane region
Family:Magnesium transport protein CorA, transmembrane region |
| 33 | c2y69Z_ |
|
not modelled |
6.7 |
24 |
PDB header:electron transport
Chain: Z: PDB Molecule:cytochrome c oxidase polypeptide 8h;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
|
| 34 | c1qo8A_ |
|
not modelled |
6.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavocytochrome c3 fumarate reductase;
PDBTitle: the structure of the open conformation of a flavocytochrome2 c3 fumarate reductase
|
| 35 | d1qmha2 |
|
not modelled |
6.5 |
23 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:RNA 3'-terminal phosphate cyclase, RPTC |
| 36 | c2kncA_ |
|
not modelled |
6.5 |
8 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 37 | c1qhdA_ |
|
not modelled |
6.5 |
32 |
PDB header:viral protein
Chain: A: PDB Molecule:viral capsid vp6;
PDBTitle: crystal structure of vp6, the major capsid protein of group a2 rotavirus
|
| 38 | c3kz5E_ |
|
not modelled |
6.5 |
44 |
PDB header:dna binding protein
Chain: E: PDB Molecule:protein sopb;
PDBTitle: structure of cdomain
|
| 39 | d1roca_ |
|
not modelled |
6.4 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ASF1-like
Family:ASF1-like |
| 40 | d1kp6a_ |
|
not modelled |
6.4 |
21 |
Fold:Ferredoxin-like
Superfamily:Killer toxin KP6 alpha-subunit
Family:Killer toxin KP6 alpha-subunit |
| 41 | d2joza1 |
|
not modelled |
6.4 |
15 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 42 | c3c0fB_ |
|
not modelled |
6.4 |
36 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein af_1514;
PDBTitle: crystal structure of a novel non-pfam protein af1514 from archeoglobus2 fulgidus dsm 4304 solved by s-sad using a cr x-ray source
|
| 43 | c2l9uA_ |
|
not modelled |
6.3 |
32 |
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
|
| 44 | c1bzkA_ |
|
not modelled |
6.2 |
50 |
PDB header:transport protein
Chain: A: PDB Molecule:protein (band 3 anion transport protein);
PDBTitle: structural studies on the effects of the deletion in the2 red cell anion exchanger (band3, ae1) associated with3 south east asian ovalocytosis.
|
| 45 | c1iijA_ |
|
not modelled |
6.2 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:erbb-2 receptor protein-tyrosine kinase;
PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
|
| 46 | c2embA_ |
|
not modelled |
6.1 |
36 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 473;
PDBTitle: solution structure of the c2h2 type zinc finger (region 342-2 372) of human zinc finger protein 473
|
| 47 | d1f6ga_ |
|
not modelled |
6.1 |
14 |
Fold:Voltage-gated potassium channels
Superfamily:Voltage-gated potassium channels
Family:Voltage-gated potassium channels |
| 48 | d2h8pc1 |
|
not modelled |
6.0 |
13 |
Fold:Voltage-gated potassium channels
Superfamily:Voltage-gated potassium channels
Family:Voltage-gated potassium channels |
| 49 | c3pqvD_ |
|
not modelled |
5.9 |
9 |
PDB header:unknown function
Chain: D: PDB Molecule:rcl1 protein;
PDBTitle: cyclase homolog
|
| 50 | c2k9yB_ |
|
not modelled |
5.9 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
| 51 | c2k9yA_ |
|
not modelled |
5.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
| 52 | c3nzqB_ |
|
not modelled |
5.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
|
| 53 | d2cu9a1 |
|
not modelled |
5.8 |
10 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ASF1-like
Family:ASF1-like |
| 54 | c2xzmO_ |
|
not modelled |
5.8 |
12 |
PDB header:ribosome
Chain: O: PDB Molecule:rps13e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 55 | c2zxeA_ |
|
not modelled |
5.8 |
9 |
PDB header:hydrolase/transport protein
Chain: A: PDB Molecule:na, k-atpase alpha subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
|
| 56 | d1t98a2 |
|
not modelled |
5.7 |
14 |
Fold:STAT-like
Superfamily:MukF C-terminal domain-like
Family:MukF C-terminal domain-like |
| 57 | d1f8ea_ |
|
not modelled |
5.5 |
12 |
Fold:6-bladed beta-propeller
Superfamily:Sialidases
Family:Sialidases (neuraminidases) |
| 58 | c3n2oA_ |
|
not modelled |
5.4 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
|
| 59 | c1qmiC_ |
|
not modelled |
5.4 |
23 |
PDB header:rna 3'-terminal phosphate cyclase
Chain: C: PDB Molecule:rna 3'-terminal phosphate cyclase;
PDBTitle: crystal structure of rna 3'-terminal phosphate cyclase, an2 ubiquitous enzyme with unusual topology
|
| 60 | c3imkA_ |
|
not modelled |
5.4 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative molybdenum carrier protein;
PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
|
| 61 | c1nmbN_ |
|
not modelled |
5.4 |
12 |
PDB header:complex (hydrolase/immunoglobulin)
Chain: N: PDB Molecule:n9 neuraminidase;
PDBTitle: the structure of a complex between the nc10 antibody and influenza2 virus neuraminidase and comparison with the overlapping binding site3 of the nc41 antibody
|
| 62 | c2rmhA_ |
|
not modelled |
5.4 |
30 |
PDB header:hormone
Chain: A: PDB Molecule:urocortin-3;
PDBTitle: human urocortin 3
|
| 63 | d1v54m_ |
|
not modelled |
5.3 |
24 |
Fold:Single transmembrane helix
Superfamily:Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family:Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX) |
| 64 | c2d46A_ |
|
not modelled |
5.3 |
5 |
PDB header:metal transport
Chain: A: PDB Molecule:calcium channel, voltage-dependent, beta 4
PDBTitle: solution structure of the human beta4a-a domain
|
| 65 | c1w21D_ |
|
not modelled |
5.2 |
3 |
PDB header:hydrolase
Chain: D: PDB Molecule:neuraminidase;
PDBTitle: structure of neuraminidase from english duck subtype n62 complexed with 30 mm sialic acid (nana, neu5ac), crystal3 soaked for 43 hours at 291 k.
|
| 66 | c2cpbA_ |
|
not modelled |
5.2 |
18 |
PDB header:viral protein
Chain: A: PDB Molecule:m13 major coat protein;
PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
|
| 67 | c2vdaB_ |
|
not modelled |
5.2 |
63 |
PDB header:protein transport
Chain: B: PDB Molecule:maltoporin;
PDBTitle: solution structure of the seca-signal peptide complex
|