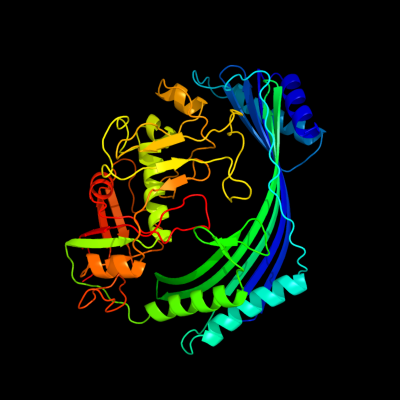
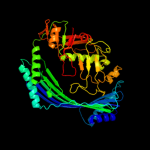
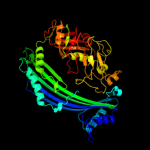
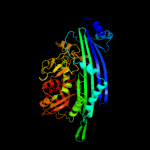
1 c3qtdC_
100.0
21
PDB header: gene regulationChain: C: PDB Molecule: pmba protein;PDBTitle: crystal structure of putative modulator of gyrase (pmba) from2 pseudomonas aeruginosa pao1
2 d1vpba_
100.0
21
Fold: Putative modulator of DNA gyrase, PmbA/TldDSuperfamily: Putative modulator of DNA gyrase, PmbA/TldDFamily: Putative modulator of DNA gyrase, PmbA/TldD3 d1vl4a_
100.0
20
Fold: Putative modulator of DNA gyrase, PmbA/TldDSuperfamily: Putative modulator of DNA gyrase, PmbA/TldDFamily: Putative modulator of DNA gyrase, PmbA/TldD4 c1bknA_
35.7
6
PDB header: dna repairChain: A: PDB Molecule: mutl;PDBTitle: crystal structure of an n-terminal 40kd fragment of e. coli2 dna mismatch repair protein mutl
5 c3kwsB_
32.4
13
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
6 d1o12a1
30.3
43
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA7 c3onrI_
30.3
0
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
8 d1vlva1
28.3
23
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase9 c1w7vD_
24.0
16
PDB header: hydrolaseChain: D: PDB Molecule: xaa-pro aminopeptidase;PDBTitle: znmg substituted aminopeptidase p from e. coli
10 d2q02a1
18.7
7
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like11 c3ff0A_
15.9
12
PDB header: biosynthetic proteinChain: A: PDB Molecule: phenazine biosynthesis protein phzb 2;PDBTitle: crystal structure of a phenazine biosynthesis-related protein (phzb2)2 from pseudomonas aeruginosa at 1.90 a resolution
12 c3oqtP_
14.9
16
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
13 d1m1ha1
14.5
28
Fold: N-utilization substance G protein NusG, insert domainSuperfamily: N-utilization substance G protein NusG, insert domainFamily: N-utilization substance G protein NusG, insert domain14 d1bkna2
14.4
6
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain15 c2iv0A_
13.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: isocitrate dehydrogenase;PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
16 c2bp1C_
13.6
14
PDB header: oxidoreductaseChain: C: PDB Molecule: aflatoxin b1 aldehyde reductase member 2;PDBTitle: structure of the aflatoxin aldehyde reductase in complex2 with nadph
17 c1ea6A_
13.3
9
PDB header: dna repairChain: A: PDB Molecule: pms1 protein homolog 2;PDBTitle: n-terminal 40kda fragment of nhpms2 complexed with adp
18 d1ydua1
13.0
18
Fold: At5g01610-likeSuperfamily: At5g01610-likeFamily: At5g01610-like19 c2qw5B_
12.4
12
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
20 d1tuha_
12.1
8
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Hypothetical protein egc068 from a soil-derived mobile gene cassette21 c1tuhA_
not modelled
12.1
8
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein egc068;PDBTitle: structure of bal32a from a soil-derived mobile gene cassette
22 c1vlvA_
not modelled
11.3
23
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
23 d1c75a_
not modelled
10.8
23
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c24 d351ca_
not modelled
10.2
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c25 d1fi3a_
not modelled
10.2
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c26 d1dvva_
not modelled
9.8
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c27 c3gknA_
not modelled
9.7
12
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein;PDBTitle: insights into the alkyl peroxide reduction activity of xanthomonas2 campestris bacterioferritin comigratory protein from the trapped3 intermediate/ligand complex structures
28 c3ezjA_
not modelled
8.9
12
PDB header: protein transportChain: A: PDB Molecule: general secretion pathway protein gspd;PDBTitle: crystal structure of the n-terminal domain of the secretin gspd from2 etec determined with the assistance of a nanobody
29 d1cora_
not modelled
8.8
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c30 c3tr9A_
not modelled
8.5
10
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
31 c3updA_
not modelled
8.4
17
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: 2.9 angstrom crystal structure of ornithine carbamoyltransferase2 (argf) from vibrio vulnificus
32 d7odca2
not modelled
8.3
10
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain33 c2on3A_
not modelled
8.2
13
PDB header: lyaseChain: A: PDB Molecule: ornithine decarboxylase;PDBTitle: a structural insight into the inhibition of human and2 leishmania donovani ornithine decarboxylases by 3-aminooxy-3 1-aminopropane
34 d1ynra1
not modelled
8.2
0
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c35 c3l23A_
not modelled
8.1
19
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
36 c2xrfA_
not modelled
8.1
13
PDB header: transferaseChain: A: PDB Molecule: uridine phosphorylase 2;PDBTitle: crystal structure of human uridine phosphorylase 2
37 d1otha1
not modelled
8.0
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase38 c3f7xA_
not modelled
8.0
19
PDB header: unknown functionChain: A: PDB Molecule: putative polyketide cyclase;PDBTitle: crystal structure of a putative polyketide cyclase (pp0894) from2 pseudomonas putida kt2440 at 1.24 a resolution
39 d1h7sa2
not modelled
8.0
9
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain40 d1b63a2
not modelled
7.9
6
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain41 d1ix2a_
not modelled
7.8
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)42 d2pjua1
not modelled
7.8
21
Fold: Chelatase-likeSuperfamily: PrpR receptor domain-likeFamily: PrpR receptor domain-like43 d2ds5a1
not modelled
7.8
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: ClpX chaperone zinc binding domain44 c1ovxB_
not modelled
7.7
20
PDB header: metal binding proteinChain: B: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: nmr structure of the e. coli clpx chaperone zinc binding domain dimer
45 c3cjlA_
not modelled
7.7
31
PDB header: unknown functionChain: A: PDB Molecule: domain of unknown function;PDBTitle: crystal structure of a protein of unknown function (eca1910) from2 pectobacterium atrosepticum scri1043 at 2.20 a resolution
46 c3ixrA_
not modelled
7.6
12
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein;PDBTitle: crystal structure of xylella fastidiosa prxq c47s mutant
47 c3h4lB_
not modelled
7.2
12
PDB header: dna binding protein, protein bindingChain: B: PDB Molecule: dna mismatch repair protein pms1;PDBTitle: crystal structure of n terminal domain of a dna repair protein
48 d1dxha1
not modelled
7.2
17
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase49 d1w0da_
not modelled
7.0
14
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases50 d2a5yb1
not modelled
7.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CED-4 C-terminal domain-like51 d2k54a1
not modelled
6.7
21
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Atu0742-like52 d1d7ka2
not modelled
6.7
10
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain53 d1pvva1
not modelled
6.7
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase54 c3na3A_
not modelled
6.6
3
PDB header: protein bindingChain: A: PDB Molecule: dna mismatch repair protein mlh1;PDBTitle: mutl protein homolog 1 isoform 1 from homo sapiens
55 c2kk4A_
not modelled
6.6
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein af_2094;PDBTitle: solution nmr structure of protein af2094 from archaeoglobus2 fulgidus. northeast structural genomics consotium (nesg)3 target gt2
56 c1htrP_
not modelled
6.5
19
PDB header: aspartyl proteaseChain: P: PDB Molecule: progastricsin (pro segment);PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
57 d1s5aa_
not modelled
6.5
13
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: PhzA/PhzB-like58 d1kfia4
not modelled
6.5
16
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain59 c3fuyC_
not modelled
6.4
19
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative integron gene cassette protein;PDBTitle: structure from the mobile metagenome of cole harbour salt2 marsh: integron cassette protein hfx_cass1
60 c3e8vA_
not modelled
6.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible transglutaminase-family protein;PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
61 d1ajza_
not modelled
6.3
12
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase62 c3k30B_
not modelled
6.3
21
PDB header: oxidoreductaseChain: B: PDB Molecule: histamine dehydrogenase;PDBTitle: histamine dehydrogenase from nocardiodes simplex
63 c2ds8A_
not modelled
6.3
20
PDB header: metal binding protein, protein bindingChain: A: PDB Molecule: atp-dependent clp protease atp-binding subunitPDBTitle: structure of the zbd-xb complex
64 d2c9qa1
not modelled
6.2
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)65 d1ud9a1
not modelled
6.2
16
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor66 c3o6cA_
not modelled
6.2
4
PDB header: transferaseChain: A: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
67 c2l4dA_
not modelled
6.2
20
PDB header: electron transportChain: A: PDB Molecule: sco1/senc family protein/cytochrome c;PDBTitle: cytochrome c domain of pp3183 protein from pseudomonas putida
68 d1dl6a_
not modelled
6.1
17
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain69 c2ideE_
not modelled
6.0
11
PDB header: biosynthetic proteinChain: E: PDB Molecule: molybdenum cofactor biosynthesis protein c;PDBTitle: crystal structure of the molybdenum cofactor biosynthesis protein c2 (ttha1789) from thermus theromophilus hb8
70 d1m5wa_
not modelled
6.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase71 c3r8wC_
not modelled
5.9
9
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-isopropylmalate dehydrogenase 2, chloroplastic;PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
72 d1pb1a_
not modelled
5.9
13
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases73 d1hqsa_
not modelled
5.9
17
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases74 d1ekxa1
not modelled
5.8
27
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase75 c2h90A_
not modelled
5.8
10
PDB header: oxidoreductaseChain: A: PDB Molecule: xenobiotic reductase a;PDBTitle: xenobiotic reductase a in complex with coumarin
76 c3jvvA_
not modelled
5.8
10
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
77 d1ad1a_
not modelled
5.7
12
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase78 c3blxM_
not modelled
5.7
14
PDB header: oxidoreductaseChain: M: PDB Molecule: isocitrate dehydrogenase [nad] subunit 1;PDBTitle: yeast isocitrate dehydrogenase (apo form)
79 c3mhsE_
not modelled
5.7
8
PDB header: hydrolase/transcription regulator/proteiChain: E: PDB Molecule: saga-associated factor 73;PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
80 d1cm7a_
not modelled
5.7
17
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases81 c3g0kA_
not modelled
5.6
15
PDB header: ca-binding proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: crystal structure of a protein of unknown function with a cystatin-2 like fold (saro_2880) from novosphingobium aromaticivorans dsm at3 1.30 a resolution
82 d1xaca_
not modelled
5.6
16
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases83 d1sjwa_
not modelled
5.6
17
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: SnoaL-like polyketide cyclase84 d1a05a_
not modelled
5.5
18
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases85 d1e42a2
not modelled
5.5
24
Fold: Subdomain of clathrin and coatomer appendage domainSuperfamily: Subdomain of clathrin and coatomer appendage domainFamily: Clathrin adaptor appendage, alpha and beta chain-specific domain86 c3gk0H_
not modelled
5.4
17
PDB header: transferaseChain: H: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
87 d1hyua4
not modelled
5.3
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: PDI-like88 c1yj7A_
not modelled
5.3
10
PDB header: protein transportChain: A: PDB Molecule: escj;PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
89 c3hz4A_
not modelled
5.3
9
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin;PDBTitle: crystal structure of thioredoxin from methanosarcina mazei
90 d1luza_
not modelled
5.3
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like91 d1cnza_
not modelled
5.3
17
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases92 d1unld_
not modelled
5.2
12
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin93 c1unhD_
not modelled
5.2
12
PDB header: cell cycleChain: D: PDB Molecule: cyclin-dependent kinase 5 activator 1;PDBTitle: structural mechanism for the inhibition of cdk5-p25 by2 roscovitine, aloisine and indirubin.
94 d1ddwa_
not modelled
5.2
13
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Enabled/VASP homology 1 domain (EVH1 domain)95 d1iz5a1
not modelled
5.2
14
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor96 c3b4oB_
not modelled
5.2
13
PDB header: biosynthetic proteinChain: B: PDB Molecule: phenazine biosynthesis protein a/b;PDBTitle: crystal structure of phenazine biosynthesis protein phza/b2 from burkholderia cepacia r18194, apo form
97 d1wgka_
not modelled
5.1
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog98 d1g2ua_
not modelled
5.1
15
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases99 d1yhta1
not modelled
5.1
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain