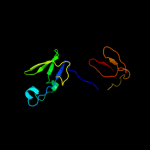
1 d1vcta2
99.5
19
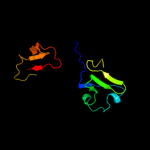
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like2 d2fy8a2
99.5
22
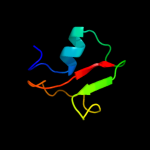
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like3 c2bknA_
99.4
19
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein ph0236;PDBTitle: structure analysis of unknown function protein
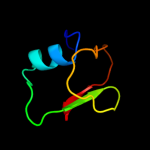
4 c3jxoB_
99.1
26
PDB header: transport proteinChain: B: PDB Molecule: trka-n domain protein;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
5 c2fy8A_
99.0
22
PDB header: transport proteinChain: A: PDB Molecule: calcium-gated potassium channel mthk;PDBTitle: crystal structure of mthk rck domain in its ligand-free gating-ring2 form
6 c1lnqC_
99.0
23
PDB header: metal transportChain: C: PDB Molecule: potassium channel related protein;PDBTitle: crystal structure of mthk at 3.3 a
7 c3l4bG_
98.3
28
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
8 c3mt5A_
97.5
11
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: potassium large conductance calcium-activated channel,PDBTitle: crystal structure of the human bk gating apparatus
9 c3u6nC_
97.3
11
PDB header: transport proteinChain: C: PDB Molecule: high-conductance ca2+-activated k+ channel protein;PDBTitle: open structure of the bk channel gating ring
10 c3nafA_
86.1
17
PDB header: ion transportChain: A: PDB Molecule: calcium-activated potassium channel subunit alpha-1;PDBTitle: structure of the intracellular gating ring from the human high-2 conductance ca2+ gated k+ channel (bk channel)
11 c2ka9A_
75.1
13
PDB header: cell adhesionChain: A: PDB Molecule: disks large homolog 4;PDBTitle: solution structure of psd-95 pdz12 complexed with cypin2 peptide
12 d1zud21
72.1
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS13 c2w8iG_
67.3
20
PDB header: membrane proteinChain: G: PDB Molecule: putative outer membrane lipoprotein wza;PDBTitle: crystal structure of wza24-345.
14 c2j58G_
64.7
20
PDB header: membrane proteinChain: G: PDB Molecule: outer membrane lipoprotein wza;PDBTitle: the structure of wza
15 d2cu3a1
64.2
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS16 c2kl0A_
63.7
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
17 c2zc3F_
58.5
9
PDB header: biosynthetic proteinChain: F: PDB Molecule: penicillin-binding protein 2x;PDBTitle: penicillin-binding protein 2x (pbp 2x) acyl-enzyme complex2 (biapenem) from streptococcus pneumoniae
18 c3cwiA_
58.3
9
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
19 d1rp5a2
57.5
15
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain20 c3r0hA_
53.9
17
PDB header: peptide binding proteinChain: A: PDB Molecule: inactivation-no-after-potential d protein;PDBTitle: structure of inad pdz45 in complex with ng2 peptide
21 d1tygb_
not modelled
53.6
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS22 d1pyya2
not modelled
51.4
13
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain23 c1tygG_
not modelled
49.6
10
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
24 d1k25a2
not modelled
48.8
13
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain25 d1k25a1
not modelled
47.8
12
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain26 d1pyya1
not modelled
45.6
8
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain27 c2anrA_
not modelled
45.1
14
PDB header: rna-binding protein/rnaChain: A: PDB Molecule: neuro-oncological ventral antigen 1;PDBTitle: crystal structure (ii) of nova-1 kh1/kh2 domain tandem with 25nt rna2 hairpin
28 d1r61a_
not modelled
34.6
11
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Putative cyclaseFamily: Putative cyclase29 c1u3bA_
not modelled
30.4
12
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding,PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
30 c2dazA_
not modelled
29.1
13
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the 7th pdz domain of inad-like2 protein
31 c2c2xB_
not modelled
27.8
10
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase-PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
32 c2qndA_
not modelled
27.3
19
PDB header: rna binding proteinChain: A: PDB Molecule: fmr1 protein;PDBTitle: crystal structure of the kh1-kh2 domains from human fragile x mental2 retardation protein
33 c3krmB_
not modelled
25.5
14
PDB header: rna binding proteinChain: B: PDB Molecule: insulin-like growth factor 2 mrna-binding proteinPDBTitle: imp1 kh34
34 d1v62a_
not modelled
23.8
15
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain35 d1wpga1
not modelled
21.5
19
Fold: Double-stranded beta-helixSuperfamily: Calcium ATPase, transduction domain AFamily: Calcium ATPase, transduction domain A36 c3tl6B_
not modelled
21.0
12
PDB header: transferaseChain: B: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of purine nucleoside phosphorylase from entamoeba2 histolytica
37 d1v6ba_
not modelled
20.6
15
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain38 c2eehA_
not modelled
19.9
12
PDB header: metal binding proteinChain: A: PDB Molecule: pdz domain-containing protein 7;PDBTitle: solution structure of first pdz domain of pdz domain2 containing protein 7
39 d1x6da1
not modelled
19.7
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: Interleukin 1640 d1sdwa2
not modelled
19.4
21
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: PHM/PNGase FFamily: Peptidylglycine alpha-hydroxylating monooxygenase, PHM41 c2hc8A_
not modelled
18.5
21
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
42 d2h1qa1
not modelled
18.2
23
Fold: PLP-dependent transferase-likeSuperfamily: Dhaf3308-likeFamily: Dhaf3308-like43 d2i0ia1
not modelled
17.4
12
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain44 c2kegA_
not modelled
17.4
22
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnk;PDBTitle: nmr structure of plantaricin k in dpc-micelles
45 c2jkdB_
not modelled
16.7
26
PDB header: gene regulationChain: B: PDB Molecule: pre-mrna leakage protein 1;PDBTitle: structure of the yeast pml1 splicing factor and its2 integration into the res complex
46 c1yj5C_
not modelled
16.2
38
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
47 d1pm3a_
not modelled
16.0
19
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: MTH189548 d1y7ma2
not modelled
15.8
24
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain49 c2jvzA_
not modelled
15.7
20
PDB header: splicingChain: A: PDB Molecule: far upstream element-binding protein 2;PDBTitle: solution nmr structure of the second and third kh domains2 of ksrp
50 d1edza1
not modelled
15.1
6
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain51 c2djtA_
not modelled
15.0
10
PDB header: signaling proteinChain: A: PDB Molecule: unnamed protein product;PDBTitle: solution structures of the pdz domain of human unnamed2 protein product
52 d1ky9a1
not modelled
14.9
15
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases53 d2ix0a3
not modelled
14.8
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like54 c2fpeB_
not modelled
14.6
12
PDB header: signaling proteinChain: B: PDB Molecule: c-jun-amino-terminal kinase interacting proteinPDBTitle: conserved dimerization of the ib1 src-homology 3 domain
55 c3kt9A_
not modelled
14.2
24
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
56 d2piaa1
not modelled
13.9
15
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like57 c2xlkB_
not modelled
13.7
16
PDB header: hydrolase/rnaChain: B: PDB Molecule: csy4 endoribonuclease;PDBTitle: crystal structure of the csy4-crrna complex, orthorhombic form
58 c2dmzA_
not modelled
13.7
9
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the third pdz domain of human inad-2 like protein
59 d1yjma1
not modelled
13.6
38
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain60 d1ujxa_
not modelled
13.5
41
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain61 d1vhwa_
not modelled
13.4
25
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases62 c2yt7A_
not modelled
13.2
10
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding familyPDBTitle: solution structure of the pdz domain of amyloid beta a42 precursor protein-binding family a member 3
63 c2gzvA_
not modelled
13.0
14
PDB header: signaling proteinChain: A: PDB Molecule: prkca-binding protein;PDBTitle: the cystal structure of the pdz domain of human pick1 (casp target)
64 c2d8iA_
not modelled
12.9
15
PDB header: immune system, signaling proteinChain: A: PDB Molecule: t-cell lymphoma invasion and metastasis 1PDBTitle: solution structure of the pdz domain of t-cell lymphoma2 invasion and metastasis 1 varian
65 c1zcdA_
not modelled
12.9
20
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: crystal structure of the na+/h+ antiporter nhaa
66 d1wgka_
not modelled
12.6
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog67 d1efna_
not modelled
12.5
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain68 c3cqtA_
not modelled
12.4
15
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fyn;PDBTitle: n53i v55l mutant of fyn sh3 domain
69 d1wi2a_
not modelled
12.4
11
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain70 d1shfa_
not modelled
12.3
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain71 c4a8aI_
not modelled
12.2
18
PDB header: hydrolase/hydrolaseChain: I: PDB Molecule: periplasmic ph-dependent serine endoprotease degq;PDBTitle: asymmetric cryo-em reconstruction of e. coli degq 12-mer in complex2 with lysozyme
72 c2y0oA_
not modelled
12.1
23
PDB header: isomeraseChain: A: PDB Molecule: probable d-lyxose ketol-isomerase;PDBTitle: the structure of a d-lyxose isomerase from the sigmab2 regulon of bacillus subtilis
73 c2k9xA_
not modelled
12.0
23
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of urm1 from trypanosoma brucei
74 d1wi4a1
not modelled
11.9
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain75 d1k9sa_
not modelled
11.8
23
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases76 c1edzA_
not modelled
11.6
6
PDB header: oxidoreductaseChain: A: PDB Molecule: 5,10-methylenetetrahydrofolate dehydrogenase;PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
77 c2kncA_
not modelled
11.6
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
78 c2kfwA_
not modelled
11.2
25
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerasePDBTitle: solution structure of full-length slyd from e.coli
79 c3elsA_
not modelled
11.1
26
PDB header: splicingChain: A: PDB Molecule: pre-mrna leakage protein 1;PDBTitle: crystal structure of yeast pml1p, residues 51-204
80 d1qcfa1
not modelled
11.1
19
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain81 d2piea1
not modelled
10.9
20
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain82 d2d6fa1
not modelled
10.9
12
Fold: Sm-like foldSuperfamily: GatD N-terminal domain-likeFamily: GatD N-terminal domain-like83 c1mhsA_
not modelled
10.8
21
PDB header: membrane protein, proton transportChain: A: PDB Molecule: plasma membrane atpase;PDBTitle: model of neurospora crassa proton atpase
84 d1lgpa_
not modelled
10.7
29
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain85 d1va8a1
not modelled
10.6
20
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain86 c2egeA_
not modelled
10.5
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein kiaa1666;PDBTitle: solution structure of the third sh3 domain from human2 kiaa1666 protein
87 c1w9qB_
not modelled
10.2
14
PDB header: cell adhesionChain: B: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the pdz tandem of human syntenin in2 complex with tnefaf peptide
88 d1viga_
not modelled
9.8
14
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)89 c2dgrA_
not modelled
9.8
24
PDB header: rna binding proteinChain: A: PDB Molecule: ring finger and kh domain-containing protein 1;PDBTitle: solution structure of the second kh domain in ring finger2 and kh domain containing protein 1
90 c2egaA_
not modelled
9.7
15
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the first sh3 domain from human2 kiaa0418 protein
91 d1ufxa_
not modelled
9.6
9
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain92 c3kw0D_
not modelled
9.5
25
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
93 d1g9oa_
not modelled
9.5
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain94 c2ejyA_
not modelled
9.5
10
PDB header: membrane proteinChain: A: PDB Molecule: 55 kda erythrocyte membrane protein;PDBTitle: solution structure of the p55 pdz t85c domain complexed2 with the glycophorin c f127c peptide
95 d1rgwa_
not modelled
9.4
12
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain96 c1tuaA_
not modelled
9.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ape0754;PDBTitle: 1.5 a crystal structure of a protein of unknown function2 ape0754 from aeropyrum pernix
97 c3npgD_
not modelled
9.2
11
PDB header: unknown functionChain: D: PDB Molecule: uncharacterized duf364 family protein;PDBTitle: crystal structure of a protein with unknown function from duf3642 family (ph1506) from pyrococcus horikoshii at 2.70 a resolution
98 c2dmoA_
not modelled
9.2
12
PDB header: signaling proteinChain: A: PDB Molecule: neutrophil cytosol factor 2;PDBTitle: refined solution structure of the 1st sh3 domain from human2 neutrophil cytosol factor 2 (ncf-2)
99 d1rzxa_
not modelled
9.1
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain