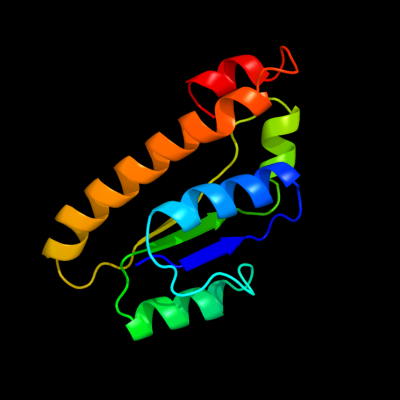
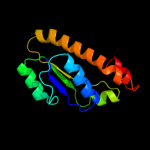
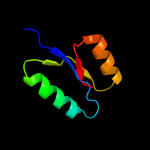
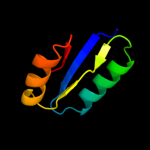
1 c3cniA_
99.4
14
PDB header: transport proteinChain: A: PDB Molecule: putative abc type-2 transporter;PDBTitle: crystal structure of a domain of a putative abc type-2 transporter2 from thermotoga maritima msb8
2 d2p0sa1
95.7
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: PG0945 N-terminal domain-like3 c3eulB_
86.3
7
PDB header: transcriptionChain: B: PDB Molecule: possible nitrate/nitrite response transcriptionalPDBTitle: structure of the signal receiver domain of the putative2 response regulator narl from mycobacterium tuberculosis
4 d1a04a2
79.2
19
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related5 c1rnlA_
77.8
19
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
6 d1a2oa1
73.1
17
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related7 c3cz5B_
70.9
15
PDB header: transcription regulatorChain: B: PDB Molecule: two-component response regulator, luxr family;PDBTitle: crystal structure of two-component response regulator, luxr family,2 from aurantimonas sp. si85-9a1
8 c1a2oB_
70.7
17
PDB header: bacterial chemotaxisChain: B: PDB Molecule: cheb methylesterase;PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
9 d1dz3a_
70.0
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related10 c3b2nA_
66.5
12
PDB header: transcriptionChain: A: PDB Molecule: uncharacterized protein q99uf4;PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
11 c3t8yA_
61.0
13
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis response regulator protein-glutamatePDBTitle: crystal structure of the response regulator domain of thermotoga2 maritima cheb
12 c2qv0A_
59.8
15
PDB header: transcriptionChain: A: PDB Molecule: protein mrke;PDBTitle: crystal structure of the response regulatory domain of2 protein mrke from klebsiella pneumoniae
13 c3c3wB_
55.2
13
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
14 c3cnbC_
52.7
21
PDB header: dna binding proteinChain: C: PDB Molecule: dna-binding response regulator, merr family;PDBTitle: crystal structure of signal receiver domain of dna binding response2 regulator protein (merr) from colwellia psychrerythraea 34h
15 d1v95a_
50.6
5
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS16 d1heya_
46.0
4
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related17 c3fkqA_
46.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ntrc-like two-domain protein;PDBTitle: crystal structure of ntrc-like two-domain protein (rer070207001320)2 from eubacterium rectale at 2.10 a resolution
18 c2gwrA_
44.4
11
PDB header: signaling proteinChain: A: PDB Molecule: dna-binding response regulator mtra;PDBTitle: crystal structure of the response regulator protein mtra from2 mycobacterium tuberculosis
19 c3f6cB_
40.9
10
PDB header: dna binding proteinChain: B: PDB Molecule: positive transcription regulator evga;PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
20 c3cg4A_
39.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-like);PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
21 c3kyiB_
not modelled
39.7
17
PDB header: transferaseChain: B: PDB Molecule: chey6 protein;PDBTitle: crystal structure of the phosphorylated p1 domain of chea3 in complex2 with chey6 from r. sphaeroides
22 d1p6qa_
not modelled
38.2
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related23 d1jbea_
not modelled
37.8
5
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related24 c3r0jA_
not modelled
33.5
11
PDB header: dna binding proteinChain: A: PDB Molecule: possible two component system response transcriptionalPDBTitle: structure of phop from mycobacterium tuberculosis
25 d2ayxa1
not modelled
32.5
7
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related26 c3k2dA_
not modelled
29.5
4
PDB header: immune systemChain: A: PDB Molecule: abc-type metal ion transport system, periplasmic component;PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
27 c1p99A_
not modelled
28.8
5
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pg110;PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
28 d1p99a_
not modelled
28.8
5
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like29 d1xs5a_
not modelled
28.8
4
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like30 d1dbwa_
not modelled
26.6
17
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related31 d1s8na_
not modelled
26.5
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related32 c1kgsA_
not modelled
26.5
14
PDB header: dna binding proteinChain: A: PDB Molecule: dna binding response regulator d;PDBTitle: crystal structure at 1.50 a of an ompr/phob homolog from thermotoga2 maritima
33 d1u0sy_
not modelled
23.9
8
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related34 c3a0rB_
not modelled
21.6
12
PDB header: transferaseChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
35 c3nhzA_
not modelled
21.5
14
PDB header: dna binding proteinChain: A: PDB Molecule: two component system transcriptional regulator mtra;PDBTitle: structure of n-terminal domain of mtra
36 c3grcD_
not modelled
20.1
13
PDB header: transferaseChain: D: PDB Molecule: sensor protein, kinase;PDBTitle: crystal structure of a sensor protein from polaromonas sp.2 js666
37 d1zgza1
not modelled
20.1
22
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related38 c3khtA_
not modelled
17.8
12
PDB header: signaling proteinChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator from hahella chejuensis
39 c3t6kB_
not modelled
17.8
19
PDB header: signaling proteinChain: B: PDB Molecule: response regulator receiver;PDBTitle: crystal structure of a hypothetical response regulator (caur_3799)2 from chloroflexus aurantiacus j-10-fl at 1.86 a resolution
40 c1zn2A_
not modelled
17.7
15
PDB header: transcription regulatorChain: A: PDB Molecule: response regulatory protein;PDBTitle: low resolution structure of response regulator styr
41 d1ys7a2
not modelled
17.4
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related42 c3chgB_
not modelled
16.5
12
PDB header: ligand binding proteinChain: B: PDB Molecule: glycine betaine-binding protein;PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
43 d1kgsa2
not modelled
16.5
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related44 c1ys7B_
not modelled
16.4
12
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulatory protein prra;PDBTitle: crystal structure of the response regulator protein prra comlexed with2 mg2+
45 c3q9sA_
not modelled
16.1
16
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding response regulator;PDBTitle: crystal structure of rra(1-215) from deinococcus radiodurans
46 c2rejA_
not modelled
16.1
3
PDB header: choline-binding proteinChain: A: PDB Molecule: putative glycine betaine abc transporter protein;PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
47 d1mvoa_
not modelled
16.0
24
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related48 d1xhfa1
not modelled
15.4
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related49 c2oqrA_
not modelled
15.3
14
PDB header: transcription,signaling proteinChain: A: PDB Molecule: sensory transduction protein regx3;PDBTitle: the structure of the response regulator regx3 from mycobacterium2 tuberculosis
50 c3hdgE_
not modelled
15.3
14
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the n-terminal domain of an2 uncharacterized protein (ws1339) from wolinella3 succinogenes
51 c2h9qC_
not modelled
15.3
9
PDB header: transcriptionChain: C: PDB Molecule: hth-type transcriptional regulator catm;PDBTitle: crystal structure of the effector binding domain of a catm2 variant (r156h)
52 c3nhmA_
not modelled
15.3
10
PDB header: signaling proteinChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure of a response regulator from myxococcus xanthus
53 c3ir1F_
not modelled
15.1
4
PDB header: protein bindingChain: F: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
54 c3breA_
not modelled
14.8
12
PDB header: signaling proteinChain: A: PDB Molecule: probable two-component response regulator;PDBTitle: crystal structure of p.aeruginosa pa3702
55 c3hv2B_
not modelled
14.4
12
PDB header: signaling proteinChain: B: PDB Molecule: response regulator/hd domain protein;PDBTitle: crystal structure of signal receiver domain of hd domain-2 containing protein from pseudomonas fluorescens pf-5
56 c3cg0A_
not modelled
13.7
6
PDB header: lyaseChain: A: PDB Molecule: response regulator receiver modulated diguanylate cyclasePDBTitle: crystal structure of signal receiver domain of modulated diguanylate2 cyclase from desulfovibrio desulfuricans g20, an example of alternate3 folding
57 c3cu5B_
not modelled
13.0
13
PDB header: transcription regulatorChain: B: PDB Molecule: two component transcriptional regulator, arac family;PDBTitle: crystal structure of a two component transcriptional regulator arac2 from clostridium phytofermentans isdg
58 c3ho7A_
not modelled
13.0
3
PDB header: transcriptionChain: A: PDB Molecule: oxyr;PDBTitle: crystal structure of oxyr from porphyromonas gingivalis
59 c2qzjC_
not modelled
13.0
18
PDB header: transcriptionChain: C: PDB Molecule: two-component response regulator;PDBTitle: crystal structure of a two-component response regulator from2 clostridium difficile
60 d1peya_
not modelled
12.9
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related61 c3l6gA_
not modelled
12.6
11
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
62 c3i42A_
not modelled
12.6
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-PDBTitle: structure of response regulator receiver domain (chey-like)2 from methylobacillus flagellatus
63 c3lteH_
not modelled
12.1
9
PDB header: transcriptionChain: H: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator (signal receiver domain) from2 bermanella marisrubri
64 c2rjnA_
not modelled
11.6
13
PDB header: hydrolaseChain: A: PDB Molecule: response regulator receiver:metal-dependentPDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
65 d1zesa1
not modelled
11.3
8
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related66 d1ny5a1
not modelled
11.1
6
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related67 c3hzhA_
not modelled
10.2
9
PDB header: signaling proteinChain: A: PDB Molecule: chemotaxis response regulator (chey-3);PDBTitle: crystal structure of the chex-chey-bef3-mg+2 complex from2 borrelia burgdorferi
68 d1yioa2
not modelled
10.0
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related69 c2qxyB_
not modelled
9.6
16
PDB header: transcriptionChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of a response regulator from thermotoga2 maritima
70 c2hqrA_
not modelled
9.6
12
PDB header: signaling proteinChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
71 d1krwa_
not modelled
9.3
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related72 c3dzdA_
not modelled
9.2
16
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4 in the inactive2 state
73 d1w25a1
not modelled
8.8
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related74 c3c3mA_
not modelled
8.7
11
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
75 c2qr3A_
not modelled
8.7
18
PDB header: transcriptionChain: A: PDB Molecule: two-component system response regulator;PDBTitle: crystal structure of the n-terminal signal receiver domain of two-2 component system response regulator from bacteroides fragilis
76 d2pl1a1
not modelled
8.7
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related77 d1zh2a1
not modelled
8.4
17
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related78 c2ep5B_
not modelled
8.4
13
PDB header: oxidoreductaseChain: B: PDB Molecule: 350aa long hypothetical aspartate-semialdehydePDBTitle: structural study of project id st1242 from sulfolobus tokodaii strain7
79 c2zwmA_
not modelled
8.2
10
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein yycf;PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
80 d2a9pa1
not modelled
8.2
10
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related81 c2f7cA_
not modelled
8.0
8
PDB header: gene regulationChain: A: PDB Molecule: hth-type transcriptional regulator catm;PDBTitle: catm effector binding domain with its effector cis,cis-muconate
82 c3muqB_
not modelled
7.9
8
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
83 d2czla1
not modelled
7.8
8
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like84 c2jrlA_
not modelled
7.8
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
85 c3c97A_
not modelled
7.5
2
PDB header: signaling protein, transferaseChain: A: PDB Molecule: signal transduction histidine kinase;PDBTitle: crystal structure of the response regulator receiver domain2 of a signal transduction histidine kinase from aspergillus3 oryzae
86 d1mb3a_
not modelled
7.4
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related87 c1ny5A_
not modelled
7.2
10
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigm54 activator (aaa+ atpase) in the inactive2 state
88 c2nt3A_
not modelled
7.2
20
PDB header: signaling proteinChain: A: PDB Molecule: response regulator homolog;PDBTitle: receiver domain from myxococcus xanthus social motility protein frzs2 (y102a mutant)
89 c2zayA_
not modelled
7.2
6
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator from desulfuromonas2 acetoxidans
90 c3tmgA_
not modelled
7.0
7
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine, l-proline abc transporter,PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
91 c3kn3C_
not modelled
6.8
2
PDB header: transcriptionChain: C: PDB Molecule: putative periplasmic protein;PDBTitle: crystal structure of lysr substrate binding domain (25-263) of2 putative periplasmic protein from wolinella succinogenes
92 c2v25B_
not modelled
6.6
5
PDB header: receptorChain: B: PDB Molecule: major cell-binding factor;PDBTitle: structure of the campylobacter jejuni antigen peb1a, an2 aspartate and glutamate receptor with bound aspartate
93 c1w25B_
not modelled
6.4
12
PDB header: signaling proteinChain: B: PDB Molecule: stalked-cell differentiation controlling protein;PDBTitle: response regulator pled in complex with c-digmp
94 c3gxaA_
not modelled
6.4
9
PDB header: protein bindingChain: A: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of gna1946
95 d1k66a_
not modelled
6.4
8
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related96 c2qsjB_
not modelled
6.4
8
PDB header: transcriptionChain: B: PDB Molecule: dna-binding response regulator, luxr family;PDBTitle: crystal structure of a luxr family dna-binding response2 regulator from silicibacter pomeroyi
97 c3cfyA_
not modelled
6.3
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative luxo repressor protein;PDBTitle: crystal structure of signal receiver domain of putative luxo2 repressor protein from vibrio parahaemolyticus
98 c3snkA_
not modelled
6.2
8
PDB header: signaling proteinChain: A: PDB Molecule: response regulator chey-like protein;PDBTitle: crystal structure of a response regulator chey-like protein (mll6475)2 from mesorhizobium loti at 2.02 a resolution
99 c3ktoA_
not modelled
6.1
8
PDB header: transcription regulatorChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver protein2 from pseudoalteromonas atlantica