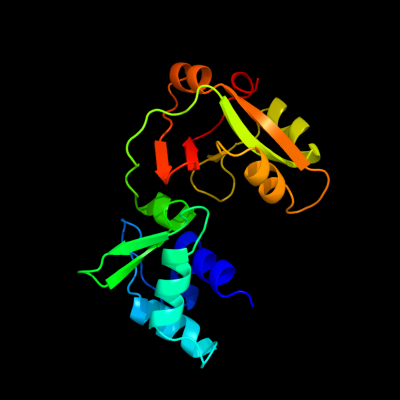
| 1 | d1wfxa_
|
|
 |
100.0 |
33 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:Tpt1/KptA |
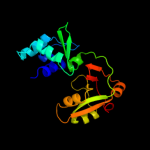
| 2 | d2cqka1
|
|
 |
96.2 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:La domain |
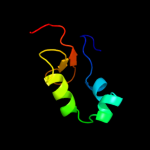
| 3 | d1s29a_
|
|
 |
96.2 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:La domain |
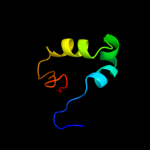
| 4 | d1zh5a1
|
|
 |
95.2 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:La domain |
| 5 | c2paxA_
|
|
 |
89.2 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:poly(adp-ribose) polymerase;
PDBTitle: the catalytic fragment of poly(adp-ribose) polymerase2 complexed with 4-amino-1,8-naphthalimide
|
| 6 | c1gs0B_
|
|
 |
87.5 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:poly (adp-ribose) polymerase-2;
PDBTitle: crystal structure of the catalytic fragment of murine poly2 (adp-ribose) polymerase-2
|
| 7 | d1gs0a2
|
|
 |
86.5 |
23 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:Poly(ADP-ribose) polymerase, C-terminal domain |
| 8 | c3c4hA_
|
|
 |
84.6 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:poly(adp-ribose) polymerase 3;
PDBTitle: human poly(adp-ribose) polymerase 3, catalytic fragment in complex2 with an inhibitor dr2313
|
| 9 | c2x5yA_
|
|
 |
81.4 |
15 |
PDB header:immune system
Chain: A: PDB Molecule:zinc finger ccch-type antiviral protein 1;
PDBTitle: human zc3hav1 (artd13), c-terminal domain
|
| 10 | c3q6nF_
|
|
 |
80.4 |
15 |
PDB header:chaperone
Chain: F: PDB Molecule:heat shock protein hsp 90-alpha;
PDBTitle: crystal structure of human mc-hsp90 in p21 space group
|
| 11 | d1efya2
|
|
 |
78.8 |
23 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:Poly(ADP-ribose) polymerase, C-terminal domain |
| 12 | c2vopA_
|
|
 |
78.7 |
15 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:lupus la protein;
PDBTitle: crystal structure of n-terminal domains of human la protein2 complexed with rna oligomer auuuu
|
| 13 | c3goyD_
|
|
 |
74.6 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:poly [adp-ribose] polymerase 14;
PDBTitle: crystal structure of human poly(adp-ribose) polymerase 14,2 catalytic fragment in complex with an inhibitor 3-3 aminobenzamide
|
| 14 | d2rd6a2
|
|
 |
74.4 |
23 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:Poly(ADP-ribose) polymerase, C-terminal domain |
| 15 | c3bljA_
|
|
 |
73.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:poly(adp-ribose) polymerase 15;
PDBTitle: crystal structure of human poly(adp-ribose) polymerase 15, catalytic2 fragment
|
| 16 | c2pqfF_
|
|
 |
71.3 |
36 |
PDB header:transferase
Chain: F: PDB Molecule:poly [adp-ribose] polymerase 12;
PDBTitle: human poly(adp-ribose) polymerase 12, catalytic fragment in complex2 with an inhibitor 3-aminobenzoic acid
|
| 17 | c3pryA_
|
|
 |
66.9 |
17 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hsp 90-beta;
PDBTitle: crystal structure of the middle domain of human hsp90-beta refined at2 2.3 a resolution
|
| 18 | c3hkvA_
|
|
 |
63.4 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:poly [adp-ribose] polymerase 10;
PDBTitle: human poly(adp-ribose) polymerase 10, catalytic fragment in2 complex with an inhibitor 3-aminobenzamide
|
| 19 | d2o0qa1
|
|
 |
62.9 |
27 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:CC0527-like |
| 20 | c1y6zA_
|
|
 |
53.1 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:heat shock protein, putative;
PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
|
| 21 | c3mhkA_ |
|
not modelled |
48.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:tankyrase-2;
PDBTitle: human tankyrase 2 - catalytic parp domain in complex with 2-(2-2 pyridyl)-7,8-dihydro-5h-thiino[4,3-d]pyrimidin-4-ol
|
| 22 | c2d58A_ |
|
not modelled |
47.1 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:allograft inflammatory factor 1;
PDBTitle: human microglia-specific protein iba1
|
| 23 | d1n0yb_ |
|
not modelled |
44.5 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 24 | c2amiA_ |
|
not modelled |
42.9 |
18 |
PDB header:cell cycle
Chain: A: PDB Molecule:caltractin;
PDBTitle: solution structure of the calcium-loaded n-terminal sensor2 domain of centrin
|
| 25 | c2vtgA_ |
|
not modelled |
40.4 |
18 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:ionized calcium-binding adapter molecule 2;
PDBTitle: crystal structure of human iba2, trigonal crystal form
|
| 26 | d1wrka1 |
|
not modelled |
40.3 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 27 | c2gq0B_ |
|
not modelled |
38.8 |
19 |
PDB header:chaperone, hydrolase
Chain: B: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of the middle domain of htpg, the e. coli2 hsp90
|
| 28 | d1sw8a_ |
|
not modelled |
38.6 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 29 | c2kzvA_ |
|
not modelled |
38.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of cv_0373(175-257) protein from2 chromobacterium violaceum, northeast structural genomics consortium3 target cvr118a
|
| 30 | c2aucC_ |
|
not modelled |
37.2 |
14 |
PDB header:membrane protein
Chain: C: PDB Molecule:myosin a tail interacting protein;
PDBTitle: structure of the plasmodium mtip-myoa complex, a key component of the2 parasite invasion motor
|
| 31 | c3hjcA_ |
|
not modelled |
36.3 |
15 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 83-1;
PDBTitle: crystal structure of the carboxy-terminal domain of hsp90 from2 leishmania major, lmjf33.0312
|
| 32 | c1hk7A_ |
|
not modelled |
35.7 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hsp82;
PDBTitle: middle domain of hsp90
|
| 33 | c2o1uA_ |
|
not modelled |
35.2 |
28 |
PDB header:chaperone
Chain: A: PDB Molecule:endoplasmin;
PDBTitle: structure of full length grp94 with amp-pnp bound
|
| 34 | d2fcea1 |
|
not modelled |
35.2 |
18 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 35 | d1tiza_ |
|
not modelled |
34.4 |
13 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 36 | c2g2bA_ |
|
not modelled |
34.4 |
21 |
PDB header:immune system
Chain: A: PDB Molecule:allograft inflammatory factor 1;
PDBTitle: nmr structure of the human allograft inflammatory factor 1
|
| 37 | c2cgeD_ |
|
not modelled |
34.2 |
20 |
PDB header:chaperone
Chain: D: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
|
| 38 | d1m39a_ |
|
not modelled |
32.8 |
24 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 39 | c2cg9A_ |
|
not modelled |
32.8 |
19 |
PDB header:chaperone
Chain: A: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
|
| 40 | c2fcdA_ |
|
not modelled |
32.7 |
19 |
PDB header:cell cycle
Chain: A: PDB Molecule:myosin light chain 1;
PDBTitle: solution structure of n-lobe myosin light chain from2 saccharomices cerevisiae
|
| 41 | d1s6ja_ |
|
not modelled |
29.7 |
18 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 42 | d1avsa_ |
|
not modelled |
29.3 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 43 | c2o1tB_ |
|
not modelled |
28.9 |
24 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin;
PDBTitle: structure of middle plus c-terminal domains (m+c) of grp94
|
| 44 | d1usua_ |
|
not modelled |
28.8 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Hsp90 middle domain |
| 45 | d1fw4a_ |
|
not modelled |
28.5 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 46 | c2o1wB_ |
|
not modelled |
27.8 |
28 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin;
PDBTitle: structure of n-terminal plus middle domains (n+m) of grp94
|
| 47 | d1ap4a_ |
|
not modelled |
27.8 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 48 | d1jc2a_ |
|
not modelled |
27.4 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 49 | c1jc2A_ |
|
not modelled |
27.4 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, skeletal muscle;
PDBTitle: complex of the c-domain of troponin c with residues 1-40 of2 troponin i
|
| 50 | d2pq3a1 |
|
not modelled |
26.6 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 51 | c2l98A_ |
|
not modelled |
26.4 |
24 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: structure of trans-resveratrol in complex with the cardiac regulatory2 protein troponin c
|
| 52 | c2kdhA_ |
|
not modelled |
26.4 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: the solution structure of human cardiac troponin c in2 complex with the green tea polyphenol; (-)-3 epigallocatechin-3-gallate
|
| 53 | c2k2aA_ |
|
not modelled |
26.1 |
16 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c;
PDBTitle: solution structure of the apo c terminal domain of lethocerus troponin2 c isoform f1
|
| 54 | d1r2ua_ |
|
not modelled |
25.2 |
18 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 55 | d1oqpa_ |
|
not modelled |
25.2 |
29 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 56 | c2jojA_ |
|
not modelled |
24.6 |
13 |
PDB header:cell cycle
Chain: A: PDB Molecule:centrin protein;
PDBTitle: nmr solution structure of n-terminal domain of euplotes2 octocarinatus centrin
|
| 57 | c2kn2A_ |
|
not modelled |
24.0 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin;
PDBTitle: solution structure of the c-terminal domain of soybean calmodulin2 isoform 4 fused with the calmodulin-binding domain of ntmkp1
|
| 58 | d1ikpa2 |
|
not modelled |
23.2 |
32 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:ADP-ribosylating toxins |
| 59 | c1ih0A_ |
|
not modelled |
22.6 |
22 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: structure of the c-domain of human cardiac troponin c in2 complex with ca2+ sensitizer emd 57033
|
| 60 | d1ih0a_ |
|
not modelled |
22.6 |
22 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 61 | c2k7bA_ |
|
not modelled |
21.7 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein 1;
PDBTitle: nmr structure of mg2+-bound cabp1 n-domain
|
| 62 | c2a4jA_ |
|
not modelled |
20.8 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:centrin 2;
PDBTitle: solution structure of the c-terminal domain (t94-y172) of2 the human centrin 2 in complex with a 17 residues peptide3 (p1-xpc) from xeroderma pigmentosum group c protein
|
| 63 | d1f54a_ |
|
not modelled |
20.4 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 64 | c3li6A_ |
|
not modelled |
19.9 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 65 | c3li6G_ |
|
not modelled |
19.9 |
13 |
PDB header:metal binding protein
Chain: G: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 66 | c3li6J_ |
|
not modelled |
19.9 |
13 |
PDB header:metal binding protein
Chain: J: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 67 | d2cxaa1 |
|
not modelled |
18.8 |
13 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:LFTR-like |
| 68 | c2cxaA_ |
|
not modelled |
18.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:leucyl/phenylalanyl-trna-protein transferase;
PDBTitle: crystal structure of leucyl/phenylalanyl-trna protein2 transferase from escherichia coli
|
| 69 | c2rf5A_ |
|
not modelled |
18.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:tankyrase-1;
PDBTitle: crystal structure of human tankyrase 1- catalytic parp domain
|
| 70 | c1scvA_ |
|
not modelled |
18.4 |
21 |
PDB header:contractile protein, structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i
|
| 71 | d1fi5a_ |
|
not modelled |
18.4 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 72 | c1sbjA_ |
|
not modelled |
18.4 |
21 |
PDB header:contractile protein, structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: nmr structure of the mg2+-loaded c terminal domain of2 cardiac troponin c bound to the n terminal domain of3 cardiac troponin i
|
| 73 | c1fi5A_ |
|
not modelled |
18.4 |
21 |
PDB header:contractile protein
Chain: A: PDB Molecule:protein (troponin c);
PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i.
|
| 74 | c3rhgA_ |
|
not modelled |
18.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phophotriesterase;
PDBTitle: crystal structure of amidohydrolase pmi1525 (target efi-500319) from2 proteus mirabilis hi4320
|
| 75 | c2kz2A_ |
|
not modelled |
17.1 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin;
PDBTitle: calmodulin, c-terminal domain, f92e mutant
|
| 76 | d1np8a_ |
|
not modelled |
15.1 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Penta-EF-hand proteins |
| 77 | c1ozsA_ |
|
not modelled |
15.0 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: c-domain of human cardiac troponin c in complex with the2 inhibitory region of human cardiac troponin i
|
| 78 | c2k9lA_ |
|
not modelled |
14.9 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpon;
PDBTitle: structure of the core binding domain of sigma54
|
| 79 | c2ktgA_ |
|
not modelled |
14.7 |
18 |
PDB header:ca-binding protein
Chain: A: PDB Molecule:calmodulin, putative;
PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein
|
| 80 | c2iopD_ |
|
not modelled |
14.2 |
17 |
PDB header:chaperone
Chain: D: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of full-length htpg, the escherichia coli2 hsp90, bound to adp
|
| 81 | d1n0ya_ |
|
not modelled |
14.1 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 82 | d1ctda_ |
|
not modelled |
13.9 |
26 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 83 | c2b1uA_ |
|
not modelled |
13.3 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin-like protein 5;
PDBTitle: solution structure of calmodulin-like skin protein c2 terminal domain
|
| 84 | d1zaca_ |
|
not modelled |
13.3 |
21 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 85 | d1cmga_ |
|
not modelled |
12.7 |
22 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 86 | c3sibA_ |
|
not modelled |
12.4 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ure3-bp sequence specific dna binding protein;
PDBTitle: crystal structure of ure3-binding protein, wild-type
|
| 87 | c1y4sA_ |
|
not modelled |
12.2 |
17 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein htpg;
PDBTitle: conformation rearrangement of heat shock protein 90 upon2 adp binding
|
| 88 | c3li6D_ |
|
not modelled |
11.9 |
13 |
PDB header:metal binding protein
Chain: D: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 89 | c2k7cA_ |
|
not modelled |
11.8 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein 1;
PDBTitle: nmr structure of mg2+-bound cabp1 c-domain
|
| 90 | d2fsua1 |
|
not modelled |
11.2 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PhnH-like
Family:PhnH-like |
| 91 | c2fsuA_ |
|
not modelled |
11.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein phnh;
PDBTitle: crystal structure of the phnh protein from escherichia coli
|
| 92 | c1npqA_ |
|
not modelled |
11.0 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c;
PDBTitle: structure of a rhodamine-labeled n-domain troponin c mutant2 (ca2+ saturated) in complex with skeletal troponin i 115-3 131
|
| 93 | c3ctnA_ |
|
not modelled |
11.0 |
21 |
PDB header:calcium-binding protein
Chain: A: PDB Molecule:troponin c;
PDBTitle: structure of calcium-saturated cardiac troponin c, nmr, 302 structures
|
| 94 | d1q46a1 |
|
not modelled |
11.0 |
37 |
Fold:SAM domain-like
Superfamily:eIF2alpha middle domain-like
Family:eIF2alpha middle domain-like |
| 95 | c1novE_ |
|
not modelled |
10.4 |
33 |
PDB header:virus
Chain: E: PDB Molecule:nodamura virus coat proteins;
PDBTitle: nodamura virus
|
| 96 | c3uatA_ |
|
not modelled |
10.3 |
13 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:disks large homolog 1;
PDBTitle: guanylate kinase domains of the maguk family scaffold proteins as2 specific phospho-protein binding modules
|
| 97 | d2qtva4 |
|
not modelled |
9.8 |
33 |
Fold:Gelsolin-like
Superfamily:C-terminal, gelsolin-like domain of Sec23/24
Family:C-terminal, gelsolin-like domain of Sec23/24 |
| 98 | d1biaa1 |
|
not modelled |
9.6 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
| 99 | c1ikqA_ |
|
not modelled |
9.5 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:exotoxin a;
PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
|