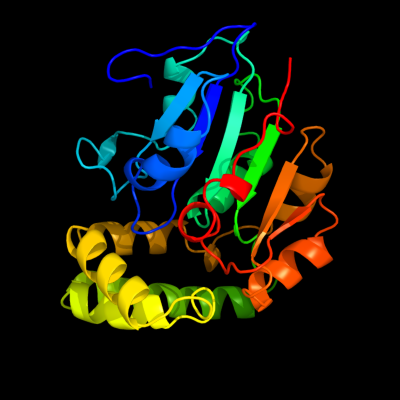
1 c1cr6A_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
2 c2qmqA_
100.0
12
PDB header: signaling proteinChain: A: PDB Molecule: protein ndrg2;PDBTitle: crystal structure of a n-myc downstream regulated 2 protein (ndrg2,2 syld, ndr2, ai182517, au040374) from mus musculus at 1.70 a3 resolution
3 c2xmzA_
100.0
27
PDB header: lyaseChain: A: PDB Molecule: hydrolase, alpha/beta hydrolase fold family;PDBTitle: structure of menh from s. aureus
4 d1r3da_
100.0
35
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hypothetical protein VC19745 c2y6vB_
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: peroxisomal membrane protein lpx1;PDBTitle: peroxisomal alpha-beta-hydrolase lpx1 (yor084w) from2 saccharomyces cerevisiae (crystal form i)
6 c2vavL_
100.0
13
PDB header: transferaseChain: L: PDB Molecule: acetyl-coa--deacetylcephalosporin cPDBTitle: crystal structure of deacetylcephalosporin c2 acetyltransferase (dac-soak)
7 c3ibtA_
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase;PDBTitle: structure of 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (qdo)
8 c3v48B_
100.0
18
PDB header: hydrolaseChain: B: PDB Molecule: putative aminoacrylate hydrolase rutd;PDBTitle: crystal structure of the putative alpha/beta hydrolase rutd from2 e.coli
9 c2wj4B_
100.0
13
PDB header: oxidoreductaseChain: B: PDB Molecule: 1h-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase;PDBTitle: crystal structure of the cofactor-devoid 1-h-3-hydroxy-4-2 oxoquinaldine 2,4-dioxygenase (hod) from arthrobacter3 nitroguajacolicus ru61a anaerobically complexed with its4 natural substrate 1-h-3-hydroxy-4-oxoquinaldine
10 c3i1iA_
100.0
15
PDB header: transferaseChain: A: PDB Molecule: homoserine o-acetyltransferase;PDBTitle: x-ray crystal structure of homoserine o-acetyltransferase from2 bacillus anthracis
11 d2b61a1
100.0
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase12 d1m33a_
100.0
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Biotin biosynthesis protein BioH13 c2r11D_
100.0
15
PDB header: hydrolaseChain: D: PDB Molecule: carboxylesterase np;PDBTitle: crystal structure of putative hydrolase (2632844) from2 bacillus subtilis at 1.96 a resolution
14 c3om8A_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: probable hydrolase;PDBTitle: the crystal structure of a hydrolase from pseudomonas aeruginosa pa01
15 c3oosA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase family protein;PDBTitle: the structure of an alpha/beta fold family hydrolase from bacillus2 anthracis str. sterne
16 c2cjpA_
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: structure of potato (solanum tuberosum) epoxide hydrolase i2 (steh1)
17 c2yysA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: proline iminopeptidase-related protein;PDBTitle: crystal structure of the proline iminopeptidase-related protein2 ttha1809 from thermus thermophilus hb8
18 c2e3jA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase ephb;PDBTitle: the crystal structure of epoxide hydrolase b (rv1938) from2 mycobacterium tuberculosis at 2.1 angstrom
19 c3qvmA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: olei00960;PDBTitle: the structure of olei00960, a hydrolase from oleispira antarctica
20 d1ehya_
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase21 d1b6ga_
not modelled
100.0
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase22 d2vata1
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase23 c2xuaH_
not modelled
100.0
16
PDB header: hydrolaseChain: H: PDB Molecule: 3-oxoadipate enol-lactonase;PDBTitle: crystal structure of the enol-lactonase from burkholderia2 xenovorans lb400
24 c3e3aA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: possible peroxidase bpoc;PDBTitle: the structure of rv0554 from mycobacterium tuberculosis
25 c3bf7B_
not modelled
100.0
18
PDB header: hydrolaseChain: B: PDB Molecule: esterase ybff;PDBTitle: 1.1 resolution structure of ybff, a new esterase from2 escherichia coli: a unique substrate-binding crevice3 generated by domain arrangement
26 d1cr6a2
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase27 d1c4xa_
not modelled
100.0
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase28 c2vf2A_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoatePDBTitle: x-ray crystal structure of hsad from mycobacterium2 tuberculosis
29 d1zd3a2
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase30 c3a2nF_
not modelled
100.0
17
PDB header: hydrolaseChain: F: PDB Molecule: haloalkane dehalogenase;PDBTitle: crystal structure of dbja (wild type type ii p21)
31 c3kxpD_
not modelled
100.0
18
PDB header: hydrolaseChain: D: PDB Molecule: alpha-(n-acetylaminomethylene)succinic acidPDBTitle: crystal structure of e-2-(acetamidomethylene)succinate2 hydrolase
32 c3e0xB_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lipase-esterase related protein;PDBTitle: the crystal structure of a lipase-esterase related protein2 from clostridium acetobutylicum atcc 824
33 c3kdaB_
not modelled
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: cftr inhibitory factor (cif);PDBTitle: crystal structure of the cftr inhibitory factor cif with the h269a2 mutation
34 c3bwxA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase;PDBTitle: crystal structure of an alpha/beta hydrolase (yp_496220.1) from2 novosphingobium aromaticivorans dsm 12444 at 1.50 a resolution
35 c1zoiC_
not modelled
100.0
15
PDB header: hydrolaseChain: C: PDB Molecule: esterase;PDBTitle: crystal structure of a stereoselective esterase from2 pseudomonas putida ifo12996
36 d2rhwa1
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase37 d1q0ra_
not modelled
100.0
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Aclacinomycin methylesterase RdmC38 c1u2eA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: 2-hydroxy-6-ketonona-2,4-dienedioic acidPDBTitle: crystal structure of the c-c bond hydrolase mhpc
39 d2pl5a1
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase40 c2xt0A_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: haloalkane dehalogenase;PDBTitle: dehalogenase dppa from plesiocystis pacifica sir-i
41 d1a8sa_
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase42 d1qo7a_
not modelled
100.0
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase43 d1mtza_
not modelled
100.0
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Proline iminopeptidase-like44 c1wprA_
not modelled
100.0
18
PDB header: signaling proteinChain: A: PDB Molecule: sigma factor sigb regulation protein rsbq;PDBTitle: crystal structure of rsbq inhibited by pmsf
45 c3l80A_
not modelled
100.0
13
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein smu.1393c;PDBTitle: crystal structure of smu.1393c from streptococcus mutans ua159
46 d1bn7a_
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase47 c3u1tA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: dmma haloalkane dehalogenase;PDBTitle: haloalkane dehalogenase, dmma, of marine microbial origin
48 d1azwa_
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Proline iminopeptidase-like49 c3fsgC_
not modelled
100.0
16
PDB header: hydrolaseChain: C: PDB Molecule: alpha/beta superfamily hydrolase;PDBTitle: crystal structure of alpha/beta superfamily hydrolase from oenococcus2 oeni psu-1
50 d1hkha_
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase51 c1y37A_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: structure of fluoroacetate dehalogenase from burkholderia sp. fa1
52 d1brta_
not modelled
100.0
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase53 c3r0vA_
not modelled
100.0
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase fold protein;PDBTitle: the crystal structure of an alpha/beta hydrolase from sphaerobacter2 thermophilus dsm 20745.
54 d1va4a_
not modelled
100.0
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase55 c3fobA_
not modelled
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: bromoperoxidase;PDBTitle: crystal structure of bromoperoxidase from bacillus anthracis
56 d1a8qa_
not modelled
100.0
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase57 c3qyjB_
not modelled
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: alr0039 protein;PDBTitle: crystal structure of alr0039, a putative alpha/beta hydrolase from2 nostoc sp pcc 7120.
58 d1a88a_
not modelled
100.0
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase59 d1xkla_
not modelled
100.0
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like60 c3p2mA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: possible hydrolase;PDBTitle: crystal structure of a novel esterase rv0045c from mycobacterium2 tuberculosis
61 d1wm1a_
not modelled
100.0
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Proline iminopeptidase-like62 d1e89a_
not modelled
100.0
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like63 c3nwoA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: proline iminopeptidase;PDBTitle: crystal structure of proline iminopeptidase mycobacterium smegmatis
64 c3r3xA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: crystal structure of the fluoroacetate dehalogenase rpa1163 -2 asp110asn/bromoacetate
65 d1mj5a_
not modelled
100.0
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase66 c2pseA_
not modelled
100.0
12
PDB header: oxidoreductaseChain: A: PDB Molecule: renilla-luciferin 2-monooxygenase;PDBTitle: crystal structures of the luciferase and green fluorescent2 protein from renilla reniformis
67 c3flaB_
not modelled
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: rifr;PDBTitle: rifr - type ii thioesterase from rifamycin nrps/pks biosynthetic2 pathway - form 1
68 d3c70a1
not modelled
100.0
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like69 c3gzjB_
not modelled
100.0
14
PDB header: hydrolaseChain: B: PDB Molecule: polyneuridine-aldehyde esterase;PDBTitle: crystal structure of polyneuridine aldehyde esterase2 complexed with 16-epi-vellosimine
70 d1uk8a_
not modelled
100.0
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase71 c2qvbA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: haloalkane dehalogenase 3;PDBTitle: crystal structure of haloalkane dehalogenase rv2579 from mycobacterium2 tuberculosis
72 c3dqzB_
not modelled
100.0
13
PDB header: lyaseChain: B: PDB Molecule: alpha-hydroxynitrile lyase-like protein;PDBTitle: structure of the hydroxynitrile lyase from arabidopsis2 thaliana
73 c2ronA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: surfactin synthetase thioesterase subunit;PDBTitle: the external thioesterase of the surfactin-synthetase
74 c3jw8A_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: mgll protein;PDBTitle: crystal structure of human mono-glyceride lipase
75 c3qitB_
not modelled
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: polyketide synthase;PDBTitle: thioesterase domain from curacin biosynthetic pathway
76 c3c5wP_
not modelled
100.0
17
PDB header: hydrolaseChain: P: PDB Molecule: pp2a-specific methylesterase pme-1;PDBTitle: complex between pp2a-specific methylesterase pme-1 and pp2a core2 enzyme
77 c1j1iA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: meta cleavage compound hydrolase;PDBTitle: crystal structure of a his-tagged serine hydrolase involved2 in the carbazole degradation (carc enzyme)
78 d1j1ia_
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase79 d1hlga_
not modelled
100.0
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase80 c2ockA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: valacyclovir hydrolase;PDBTitle: crystal structure of valacyclovir hydrolase d123n mutant
81 d1xkta_
not modelled
100.0
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases82 c3hjuB_
not modelled
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: monoglyceride lipase;PDBTitle: crystal structure of human monoglyceride lipase
83 d1k8qa_
not modelled
100.0
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase84 d1imja_
not modelled
100.0
20
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Ccg1/TafII250-interacting factor B (Cib)85 d1pjaa_
not modelled
99.9
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterases86 c1pjaA_
not modelled
99.9
12
PDB header: hydrolaseChain: A: PDB Molecule: palmitoyl-protein thioesterase 2 precursor;PDBTitle: the crystal structure of palmitoyl protein thioesterase-2 reveals the2 basis for divergent substrate specificities of the two lysosomal3 thioesterases (ppt1 and ppt2)
87 c3dyvA_
not modelled
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: esterase d;PDBTitle: snapshots of esterase d from lactobacillus rhamnosus:2 insights into a rotation driven catalytic mechanism
88 c3bdiA_
not modelled
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ta0194;PDBTitle: crystal structure of predicted cib-like hydrolase (np_393672.1) from2 thermoplasma acidophilum at 1.45 a resolution
89 d1tqha_
not modelled
99.9
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase/lipase90 c2q0xA_
not modelled
99.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: alpha/beta hydrolase fold protein of unknown function
91 c2px6A_
not modelled
99.9
11
PDB header: transferaseChain: A: PDB Molecule: thioesterase domain;PDBTitle: crystal structure of the thioesterase domain of human fatty2 acid synthase inhibited by orlistat
92 c3h04A_
not modelled
99.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein with unknown function from2 staphylococcus aureus subsp. aureus mu50
93 c3lcrA_
not modelled
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: tautomycetin biosynthetic pks;PDBTitle: thioesterase from tautomycetin biosynthhetic pathway
94 c3qmwD_
not modelled
99.9
16
PDB header: hydrolaseChain: D: PDB Molecule: thioesterase;PDBTitle: redj with peg molecule bound in the active site
95 c3qm1A_
not modelled
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: cinnamoyl esterase;PDBTitle: crystal structure of the lactobacillus johnsonii cinnamoyl esterase2 lj0536 s106a mutant in complex with ethylferulate, form ii
96 c3llcA_
not modelled
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative hydrolase (yp_002548124.1) from2 agrobacterium vitis s4 at 1.80 a resolution
97 c3ilsA_
not modelled
99.9
11
PDB header: hydrolaseChain: A: PDB Molecule: aflatoxin biosynthesis polyketide synthase;PDBTitle: the thioesterase domain from pksa
98 d1ex9a_
not modelled
99.9
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase99 d1ispa_
not modelled
99.9
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase100 c2rauA_
not modelled
99.9
12
PDB header: hydrolaseChain: A: PDB Molecule: putative esterase;PDBTitle: crystal structure of a putative lipase (np_343859.1) from sulfolobus2 solfataricus at 1.85 a resolution
101 c2wtmC_
not modelled
99.9
18
PDB header: hydrolaseChain: C: PDB Molecule: est1e;PDBTitle: est1e from butyrivibrio proteoclasticus
102 c2hdwB_
not modelled
99.9
13
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein pa2218;PDBTitle: crystal structure of hypothetical protein pa2218 from pseudomonas2 aeruginosa
103 d1mo2a_
not modelled
99.9
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases104 c1mo2A_
not modelled
99.9
15
PDB header: transferaseChain: A: PDB Molecule: erythronolide synthase, modules 5 and 6;PDBTitle: thioesterase domain from 6-deoxyerythronolide synthase2 (debs te), ph 8.5
105 c2h7xA_
not modelled
99.9
18
PDB header: hydrolaseChain: A: PDB Molecule: type i polyketide synthase pikaiv;PDBTitle: pikromycin thioesterase adduct with reduced triketide2 affinity label
106 c2qs9A_
not modelled
99.9
17
PDB header: structural proteinChain: A: PDB Molecule: retinoblastoma-binding protein 9;PDBTitle: crystal structure of the human retinoblastoma-binding2 protein 9 (rbbp-9). nesg target hr2978
107 c3fcyB_
not modelled
99.9
14
PDB header: hydrolaseChain: B: PDB Molecule: xylan esterase 1;PDBTitle: crystal structure of acetyl xylan esterase 1 from2 thermoanaerobacterium sp. jw/sl ys485
108 d1l7aa_
not modelled
99.9
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetyl xylan esterase-like109 c3fleB_
not modelled
99.9
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: se_1780 protein;PDBTitle: se_1780 protein of unknown function from staphylococcus epidermidis.
110 c3mveB_
not modelled
99.9
12
PDB header: lyaseChain: B: PDB Molecule: upf0255 protein vv1_0328;PDBTitle: crystal structure of a novel pyruvate decarboxylase
111 c3fnbB_
not modelled
99.9
15
PDB header: hydrolaseChain: B: PDB Molecule: acylaminoacyl peptidase smu_737;PDBTitle: crystal structure of acylaminoacyl peptidase smu_737 from2 streptococcus mutans ua159
112 d2h7xa1
not modelled
99.9
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases113 d1ufoa_
not modelled
99.9
21
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hypothetical protein TT1662114 d1uxoa_
not modelled
99.8
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: YdeN-like115 c2jbwB_
not modelled
99.8
19
PDB header: hydrolaseChain: B: PDB Molecule: 2,6-dihydroxy-pseudo-oxynicotine hydrolase;PDBTitle: crystal structure of the 2,6-dihydroxy-pseudo-oxynicotine2 hydrolase.
116 c3lp5A_
not modelled
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: putative cell surface hydrolase;PDBTitle: the crystal structure of the putative cell surface hydrolase from2 lactobacillus plantarum wcfs1
117 d2jbwa1
not modelled
99.8
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: 2,6-dihydropseudooxynicotine hydrolase-like118 c3ksrA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: putative serine hydrolase;PDBTitle: crystal structure of a putative serine hydrolase (xcc3885) from2 xanthomonas campestris pv. campestris at 2.69 a resolution
119 d1qlwa_
not modelled
99.8
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: A novel bacterial esterase120 c2hu7A_
not modelled
99.8
19
PDB header: hydrolaseChain: A: PDB Molecule: acylamino-acid-releasing enzyme;PDBTitle: binding of inhibitors by acylaminoacyl peptidase