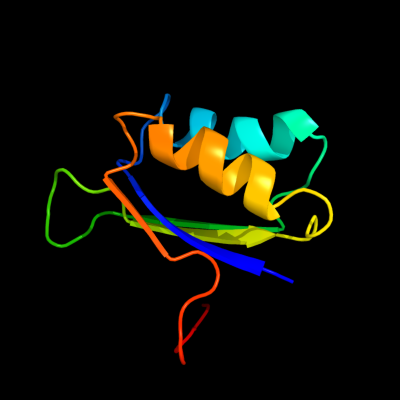
| 1 | c2pc6C_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
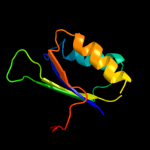
| 2 | c2fgcA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
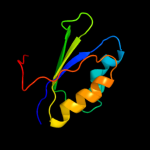
| 3 | c2f1fA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
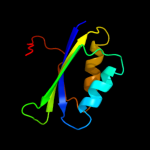
| 4 | d2fgca2
|
|
 |
100.0 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 5 | d2pc6a2
|
|
 |
99.9 |
24 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 6 | d2f1fa1
|
|
 |
99.9 |
24 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 7 | d2f06a2
|
|
 |
99.7 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 8 | d1sc6a3
|
|
 |
99.4 |
6 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 9 | d1ygya3
|
|
 |
99.3 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 10 | c2f06B_
|
|
 |
99.3 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
| 11 | c3ibwA_
|
|
 |
99.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
| 12 | c1ygyA_
|
|
 |
98.4 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
|
| 13 | d1u8sa2
|
|
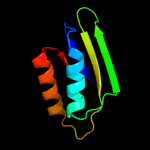 |
98.2 |
18 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 14 | c1y7pB_
|
|
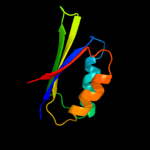 |
98.0 |
8 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein af1403;
PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
|
| 15 | d1zpva1
|
|
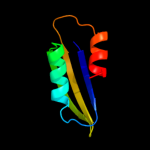 |
97.9 |
6 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 16 | d2qmwa2
|
|
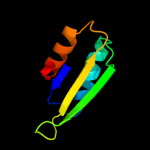 |
97.8 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 17 | c2nyiB_
|
|
 |
97.8 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 18 | d1u8sa1
|
|
 |
97.7 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 19 | c1ybaC_
|
|
 |
97.7 |
7 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: the active form of phosphoglycerate dehydrogenase
|
| 20 | c3n0vD_
|
|
 |
97.6 |
11 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 21 | c1u8sB_ |
|
not modelled |
97.6 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
| 22 | c3o1lB_ |
|
not modelled |
97.5 |
3 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 23 | c3louB_ |
|
not modelled |
97.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 24 | c3mwbA_ |
|
not modelled |
97.5 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
|
| 25 | c2qmwA_ |
|
not modelled |
97.4 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
|
| 26 | c3k5pA_ |
|
not modelled |
97.4 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
| 27 | c2qmxB_ |
|
not modelled |
97.3 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
|
| 28 | c3luyA_ |
|
not modelled |
97.3 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable chorismate mutase;
PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
|
| 29 | c3nrbD_ |
|
not modelled |
97.2 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 30 | d1phza1 |
|
not modelled |
97.2 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 31 | d2f06a1 |
|
not modelled |
97.2 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 32 | c3obiC_ |
|
not modelled |
96.9 |
8 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 33 | c3mtjA_ |
|
not modelled |
96.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
|
| 34 | c2dtjA_ |
|
not modelled |
95.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
|
| 35 | d2hmfa3 |
|
not modelled |
95.2 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 36 | d2cdqa2 |
|
not modelled |
94.1 |
21 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 37 | d2hmfa2 |
|
not modelled |
94.0 |
9 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 38 | c2re1A_ |
|
not modelled |
93.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase, alpha and beta subunits;
PDBTitle: crystal structure of aspartokinase alpha and beta subunits
|
| 39 | d2nzca1 |
|
not modelled |
92.4 |
9 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:TM1266-like |
| 40 | c2zhoB_ |
|
not modelled |
92.2 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
|
| 41 | c3mahA_ |
|
not modelled |
92.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: a putative c-terminal regulatory domain of aspartate kinase from2 porphyromonas gingivalis w83.
|
| 42 | c3mgjA_ |
|
not modelled |
90.1 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1480;
PDBTitle: crystal structure of the saccharop_dh_n domain of mj14802 protein from methanococcus jannaschii. northeast structural3 genomics consortium target mjr83a.
|
| 43 | c2dgbA_ |
|
not modelled |
89.2 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein purs;
PDBTitle: structure of thermus thermophilus purs in the p21 form
|
| 44 | c3l76B_ |
|
not modelled |
89.2 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
| 45 | d2cdqa3 |
|
not modelled |
88.0 |
21 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 46 | c2zw2B_ |
|
not modelled |
86.5 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:putative uncharacterized protein sts178;
PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
|
| 47 | d2j0wa2 |
|
not modelled |
86.4 |
21 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 48 | c1rwuA_ |
|
not modelled |
86.3 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical upf0250 protein ybed;
PDBTitle: solution structure of conserved protein ybed from e. coli
|
| 49 | d1rwua_ |
|
not modelled |
86.3 |
20 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:YbeD-like |
| 50 | c3c1nA_ |
|
not modelled |
84.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:probable aspartokinase;
PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
|
| 51 | c2phmA_ |
|
not modelled |
84.4 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (phenylalanine-4-hydroxylase);
PDBTitle: structure of phenylalanine hydroxylase dephosphorylated
|
| 52 | c3ab4K_ |
|
not modelled |
83.9 |
12 |
PDB header:transferase
Chain: K: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
|
| 53 | d1gtda_ |
|
not modelled |
83.4 |
19 |
Fold:PurS-like
Superfamily:PurS-like
Family:PurS subunit of FGAM synthetase |
| 54 | d1vq3a_ |
|
not modelled |
82.9 |
11 |
Fold:PurS-like
Superfamily:PurS-like
Family:PurS subunit of FGAM synthetase |
| 55 | c1tdjA_ |
|
not modelled |
80.9 |
17 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 56 | c2cdqB_ |
|
not modelled |
78.1 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
|
| 57 | c2rjzA_ |
|
not modelled |
76.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pilo protein;
PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
|
| 58 | d2j0wa3 |
|
not modelled |
66.3 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 59 | c3p96A_ |
|
not modelled |
65.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase serb;
PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
|
| 60 | c2jsxA_ |
|
not modelled |
62.9 |
4 |
PDB header:chaperone
Chain: A: PDB Molecule:protein napd;
PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
|
| 61 | d1t4aa_ |
|
not modelled |
56.0 |
14 |
Fold:PurS-like
Superfamily:PurS-like
Family:PurS subunit of FGAM synthetase |
| 62 | c2yx5A_ |
|
not modelled |
51.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0062 protein mj1593;
PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
|
| 63 | d1tdja2 |
|
not modelled |
42.9 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Allosteric threonine deaminase C-terminal domain |
| 64 | d2joqa1 |
|
not modelled |
42.7 |
14 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:HP0495-like |
| 65 | d1kona_ |
|
not modelled |
29.9 |
16 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
| 66 | c3ftbA_ |
|
not modelled |
28.9 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:histidinol-phosphate aminotransferase;
PDBTitle: the crystal structure of the histidinol-phosphate2 aminotransferase from clostridium acetobutylicum
|
| 67 | d1nhkl_ |
|
not modelled |
23.9 |
13 |
Fold:Ferredoxin-like
Superfamily:Nucleoside diphosphate kinase, NDK
Family:Nucleoside diphosphate kinase, NDK |
| 68 | c3cg4A_ |
|
not modelled |
23.6 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:response regulator receiver domain protein (chey-like);
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
| 69 | c2j0wA_ |
|
not modelled |
21.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:lysine-sensitive aspartokinase 3;
PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
|
| 70 | d2hoxa1 |
|
not modelled |
19.8 |
10 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 71 | d1ehwa_ |
|
not modelled |
19.4 |
27 |
Fold:Ferredoxin-like
Superfamily:Nucleoside diphosphate kinase, NDK
Family:Nucleoside diphosphate kinase, NDK |
| 72 | c3js9A_ |
|
not modelled |
19.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nucleoside diphosphate kinase family protein;
PDBTitle: crystal structure of nucleoside diphosphate kinase family protein from2 babesia bovis
|
| 73 | d2c7pa1 |
|
not modelled |
19.0 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:C5 cytosine-specific DNA methylase, DCM |
| 74 | c2hurF_ |
|
not modelled |
17.8 |
13 |
PDB header:signaling protein,transferase
Chain: F: PDB Molecule:nucleoside diphosphate kinase;
PDBTitle: escherichia coli nucleoside diphosphate kinase
|
| 75 | c3hdvB_ |
|
not modelled |
16.3 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of response regulator receiver protein from2 pseudomonas putida
|
| 76 | d1mw7a_ |
|
not modelled |
16.1 |
11 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
| 77 | d1dcta_ |
|
not modelled |
15.8 |
14 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:C5 cytosine-specific DNA methylase, DCM |
| 78 | d2b3tb1 |
|
not modelled |
15.7 |
13 |
Fold:Release factor
Superfamily:Release factor
Family:Release factor |
| 79 | d1w7wa_ |
|
not modelled |
15.6 |
12 |
Fold:Ferredoxin-like
Superfamily:Nucleoside diphosphate kinase, NDK
Family:Nucleoside diphosphate kinase, NDK |
| 80 | d1p4xa2 |
|
not modelled |
15.2 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 81 | d1qkka_ |
|
not modelled |
15.1 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 82 | d3bbba1 |
|
not modelled |
15.0 |
15 |
Fold:Ferredoxin-like
Superfamily:Nucleoside diphosphate kinase, NDK
Family:Nucleoside diphosphate kinase, NDK |
| 83 | d1lc5a_ |
|
not modelled |
14.1 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 84 | c1zbtA_ |
|
not modelled |
13.8 |
11 |
PDB header:translation
Chain: A: PDB Molecule:peptide chain release factor 1;
PDBTitle: crystal structure of peptide chain release factor 1 (rf-1) (smu.1085)2 from streptococcus mutans at 2.34 a resolution
|
| 85 | d3ctaa1 |
|
not modelled |
13.6 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 86 | d2a9pa1 |
|
not modelled |
13.5 |
8 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 87 | d1rq0a_ |
|
not modelled |
13.2 |
9 |
Fold:Release factor
Superfamily:Release factor
Family:Release factor |
| 88 | c1w7wF_ |
|
not modelled |
13.1 |
12 |
PDB header:transferase
Chain: F: PDB Molecule:nucleoside diphosphate kinase;
PDBTitle: structure and mutational analysis of a plant mitochondrial2 nucleoside diphosphate kinase: identification of residues3 involved in serine phosphorylation and oligomerization.
|
| 89 | c3me5A_ |
|
not modelled |
13.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:cytosine-specific methyltransferase;
PDBTitle: crystal structure of putative dna cytosine methylase from shigella2 flexneri 2a str. 301
|
| 90 | c3d5cX_ |
|
not modelled |
12.3 |
13 |
PDB header:ribosome
Chain: X: PDB Molecule:peptide chain release factor 1;
PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 30s subunit, release factor 1 (rf1), two trna, and3 mrna molecules of the second 70s ribosome. the entire crystal4 structure contains two 70s ribosomes as described in remark 400.
|
| 91 | c3eodA_ |
|
not modelled |
12.2 |
4 |
PDB header:signaling protein
Chain: A: PDB Molecule:protein hnr;
PDBTitle: crystal structure of n-terminal domain of e. coli rssb
|
| 92 | c2l69A_ |
|
not modelled |
12.2 |
30 |
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
|
| 93 | c3q89H_ |
|
not modelled |
12.1 |
16 |
PDB header:transferase
Chain: H: PDB Molecule:nucleoside diphosphate kinase;
PDBTitle: crystal structure of staphylococcus aureus nucleoside diphosphate2 kinase complexed with cdp
|
| 94 | c3o2sB_ |
|
not modelled |
12.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72 complex
|
| 95 | c3o2qB_ |
|
not modelled |
11.7 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
|
| 96 | d1y7pa2 |
|
not modelled |
11.5 |
6 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:AF1403 N-terminal domain-like |
| 97 | d1wxma1 |
|
not modelled |
11.1 |
7 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 98 | d1mvoa_ |
|
not modelled |
11.0 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 99 | c3fdfA_ |
|
not modelled |
10.7 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:fr253;
PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
|