| 1 | d1otja_
|
|
 |
100.0 |
100 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 2 | c3pvjB_
|
|
 |
100.0 |
61 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
|
| 3 | d1oiha_
|
|
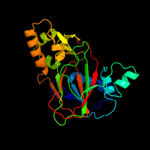 |
100.0 |
41 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 4 | c3r1jB_
|
|
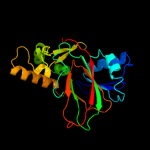 |
100.0 |
46 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
|
| 5 | c3eatX_
|
|
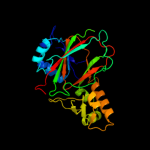 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:pyoverdine biosynthesis protein pvcb;
PDBTitle: crystal structure of the pvcb (pa2255) protein from2 pseudomonas aeruginosa
|
| 6 | d1nx4a_
|
|
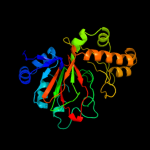 |
100.0 |
18 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 7 | c3ms5A_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gamma-butyrobetaine dioxygenase;
PDBTitle: crystal structure of human gamma-butyrobetaine,2-oxoglutarate2 dioxygenase 1 (bbox1)
|
| 8 | d1y0za_
|
|
 |
100.0 |
15 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 9 | d1ds1a_
|
|
 |
100.0 |
18 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Clavaminate synthase |
| 10 | c2og5A_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxygenase;
PDBTitle: crystal structure of asparagine oxygenase (asno)
|
| 11 | d1jr7a_
|
|
 |
100.0 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Gab protein (hypothetical protein YgaT) |
| 12 | c2wbqA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arginine beta-hydroxylase;
PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
|
| 13 | c2opwA_
|
|
 |
97.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phyhd1 protein;
PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
|
| 14 | c3nnlB_
|
|
 |
97.4 |
10 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 15 | c2rdsA_
|
|
 |
96.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxypentalenic acid 11-beta hydroxylase; fe(ii)/alpha-
PDBTitle: crystal structure of ptlh with fe/oxalylglycine and ent-1-2 deoxypentalenic acid bound
|
| 16 | d2fcta1
|
|
 |
96.5 |
14 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 17 | c3gjbA_
|
|
 |
96.1 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cytc3;
PDBTitle: cytc3 with fe(ii) and alpha-ketoglutarate
|
| 18 | c3emrA_
|
|
 |
94.9 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ectd;
PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
|
| 19 | d2a1xa1
|
|
 |
91.7 |
10 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 20 | d1wdia_
|
|
 |
85.5 |
11 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
| 21 | c1yy3A_ |
|
not modelled |
85.4 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
| 22 | d1vkya_ |
|
not modelled |
83.3 |
11 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
| 23 | c2jijA_ |
|
not modelled |
63.7 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:prolyl-4 hydroxylase;
PDBTitle: crystal structure of the apo form of chlamydomonas2 reinhardtii prolyl-4 hydroxylase type i
|
| 24 | d1gp6a_ |
|
not modelled |
59.8 |
31 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 25 | d1dcsa_ |
|
not modelled |
59.5 |
8 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 26 | c2g19A_ |
|
not modelled |
58.3 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:egl nine homolog 1;
PDBTitle: cellular oxygen sensing: crystal structure of hypoxia-2 inducible factor prolyl hydroxylase (phd2)
|
| 27 | c3pl0B_ |
|
not modelled |
56.6 |
21 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
|
| 28 | d1odma_ |
|
not modelled |
55.8 |
14 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 29 | c3itqB_ |
|
not modelled |
51.8 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prolyl 4-hydroxylase, alpha subunit domain protein;
PDBTitle: crystal structure of a prolyl 4-hydroxylase from bacillus anthracis
|
| 30 | c3ouiA_ |
|
not modelled |
49.7 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:egl nine homolog 1;
PDBTitle: phd2-r717 with 40787422
|
| 31 | c3on7C_ |
|
not modelled |
49.3 |
7 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, iron/ascorbate family;
PDBTitle: crystal structure of a putative oxygenase (so_2589) from shewanella2 oneidensis at 2.20 a resolution
|
| 32 | c2fg0B_ |
|
not modelled |
47.0 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 33 | d2csga1 |
|
not modelled |
45.8 |
12 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:YbiU-like |
| 34 | d2evra2 |
|
not modelled |
42.2 |
9 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 35 | d1e5ra_ |
|
not modelled |
40.5 |
20 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Type II Proline 3-hydroxylase (proline oxidase) |
| 36 | c2dbiA_ |
|
not modelled |
37.7 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ybiu;
PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
|
| 37 | c3gt2A_ |
|
not modelled |
34.7 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 38 | c2xivA_ |
|
not modelled |
34.2 |
19 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 39 | d1w9ya1 |
|
not modelled |
32.2 |
13 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 40 | c3npfB_ |
|
not modelled |
30.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
|
| 41 | c3pbiA_ |
|
not modelled |
28.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 42 | c3ghfA_ |
|
not modelled |
28.1 |
11 |
PDB header:cell cycle
Chain: A: PDB Molecule:septum site-determining protein minc;
PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
|
| 43 | d16pka_ |
|
not modelled |
25.9 |
20 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 44 | d1bvp12 |
|
not modelled |
25.3 |
21 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Top domain of virus capsid protein |
| 45 | c2a5hC_ |
|
not modelled |
24.9 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 46 | c1zmrA_ |
|
not modelled |
22.7 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of the e. coli phosphoglycerate kinase
|
| 47 | d1hdia_ |
|
not modelled |
20.9 |
7 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 48 | d1phpa_ |
|
not modelled |
20.0 |
14 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 49 | d1ltka_ |
|
not modelled |
19.3 |
13 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 50 | c2nnzA_ |
|
not modelled |
18.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: solution structure of the hypothetical protein af2241 from2 archaeoglobus fulgidus
|
| 51 | d1fw8a_ |
|
not modelled |
18.2 |
14 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 52 | d1vpea_ |
|
not modelled |
18.0 |
7 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 53 | c2w8iG_ |
|
not modelled |
16.9 |
31 |
PDB header:membrane protein
Chain: G: PDB Molecule:putative outer membrane lipoprotein wza;
PDBTitle: crystal structure of wza24-345.
|
| 54 | c3dkqB_ |
|
not modelled |
16.8 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pkhd-type hydroxylase sbal_3634;
PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
|
| 55 | d1v6sa_ |
|
not modelled |
16.2 |
14 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 56 | c2cunA_ |
|
not modelled |
16.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from pyrococcus2 horikoshii ot3
|
| 57 | d2toda1 |
|
not modelled |
15.7 |
8 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 58 | c3trrA_ |
|
not modelled |
15.5 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:probable enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 59 | c3q3vA_ |
|
not modelled |
14.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from campylobacter2 jejuni.
|
| 60 | d1vjda_ |
|
not modelled |
14.8 |
7 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 61 | d1qpga_ |
|
not modelled |
14.6 |
14 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 62 | c3cmwA_ |
|
not modelled |
14.6 |
22 |
PDB header:recombination/dna
Chain: A: PDB Molecule:protein reca;
PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
|
| 63 | c3h41A_ |
|
not modelled |
13.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 64 | c3bvcA_ |
|
not modelled |
13.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ism_01780;
PDBTitle: crystal structure of uncharacterized protein ism_01780 from2 roseovarius nubinhibens ism
|
| 65 | c3cmuA_ |
|
not modelled |
13.4 |
22 |
PDB header:recombination/dna
Chain: A: PDB Molecule:protein reca;
PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
|
| 66 | c2ej5B_ |
|
not modelled |
13.3 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase subunit ii;
PDBTitle: crystal structure of gk2038 protein (enoyl-coa hydratase subunit ii)2 from geobacillus kaustophilus
|
| 67 | c3rcqA_ |
|
not modelled |
13.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartyl/asparaginyl beta-hydroxylase;
PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
|
| 68 | c3r38A_ |
|
not modelled |
13.1 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.23 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase (mura) from listeria monocytogenes egd-e
|
| 69 | d1e4cp_ |
|
not modelled |
13.1 |
15 |
Fold:AraD/HMP-PK domain-like
Superfamily:AraD/HMP-PK domain-like
Family:AraD-like aldolase/epimerase |
| 70 | d7odca1 |
|
not modelled |
12.9 |
15 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 71 | c2d40C_ |
|
not modelled |
12.5 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative gentisate 1,2-dioxygenase;
PDBTitle: crystal structure of z3393 from escherichia coli o157:h7
|
| 72 | d1njjb1 |
|
not modelled |
12.4 |
8 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 73 | d2fcja1 |
|
not modelled |
12.4 |
17 |
Fold:Toprim domain
Superfamily:Toprim domain
Family:Toprim domain |
| 74 | d1zx8a1 |
|
not modelled |
12.4 |
8 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:TM1367-like |
| 75 | d1nzya_ |
|
not modelled |
11.6 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 76 | c2zpmA_ |
|
not modelled |
11.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 77 | d16vpa_ |
|
not modelled |
10.9 |
21 |
Fold:Conserved core of transcriptional regulatory protein vp16
Superfamily:Conserved core of transcriptional regulatory protein vp16
Family:Conserved core of transcriptional regulatory protein vp16 |
| 78 | c2booA_ |
|
not modelled |
10.8 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: the crystal structure of uracil-dna n-glycosylase (ung)2 from deinococcus radiodurans.
|
| 79 | d2hxma1 |
|
not modelled |
10.6 |
29 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Uracil-DNA glycosylase |
| 80 | d1ahsa_ |
|
not modelled |
10.3 |
15 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Top domain of virus capsid protein |
| 81 | d1j5ya2 |
|
not modelled |
10.3 |
5 |
Fold:HPr-like
Superfamily:Putative transcriptional regulator TM1602, C-terminal domain
Family:Putative transcriptional regulator TM1602, C-terminal domain |
| 82 | d2v9va2 |
|
not modelled |
10.2 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 83 | c2k1gA_ |
|
not modelled |
10.1 |
13 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
| 84 | d1okba_ |
|
not modelled |
10.0 |
29 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Uracil-DNA glycosylase |
| 85 | d3euga_ |
|
not modelled |
9.3 |
36 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Uracil-DNA glycosylase |
| 86 | d3bu7a1 |
|
not modelled |
9.0 |
14 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Gentisate 1,2-dioxygenase-like |
| 87 | c3bu7A_ |
|
not modelled |
9.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gentisate 1,2-dioxygenase;
PDBTitle: crystal structure and biochemical characterization of gdosp,2 a gentisate 1,2-dioxygenase from silicibacter pomeroyi
|
| 88 | c2opiB_ |
|
not modelled |
8.6 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:l-fuculose-1-phosphate aldolase;
PDBTitle: crystal structure of l-fuculose-1-phosphate aldolase from bacteroides2 thetaiotaomicron
|
| 89 | d1o98a1 |
|
not modelled |
8.5 |
38 |
Fold:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Superfamily:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain |
| 90 | c2kvcA_ |
|
not modelled |
8.5 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
|
| 91 | c2i79B_ |
|
not modelled |
8.4 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:acetyltransferase, gnat family;
PDBTitle: the crystal structure of the acetyltransferase of gnat family from2 streptococcus pneumoniae
|
| 92 | d2phda1 |
|
not modelled |
8.2 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Gentisate 1,2-dioxygenase-like |
| 93 | d1f3ta1 |
|
not modelled |
7.8 |
8 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 94 | c3ooxA_ |
|
not modelled |
7.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 2og-fe(ii) oxygenase family protein;
PDBTitle: crystal structure of a putative 2og-fe(ii) oxygenase family protein2 (cc_0200) from caulobacter crescentus at 1.44 a resolution
|
| 95 | c2zhxG_ |
|
not modelled |
7.6 |
36 |
PDB header:hydrolase/hydrolase inhibitor
Chain: G: PDB Molecule:uracil-dna glycosylase;
PDBTitle: crystal structure of uracil-dna glycosylase from mycobacterium2 tuberculosis in complex with a proteinaceous inhibitor
|
| 96 | d1dt9a3 |
|
not modelled |
7.5 |
15 |
Fold:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Superfamily:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Family:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
| 97 | d1q6za1 |
|
not modelled |
7.5 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 98 | c3rrvC_ |
|
not modelled |
7.4 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of an enoyl-coa hydratase/isomerase from2 mycobacterium paratuberculosis
|
| 99 | d1d5ra1 |
|
not modelled |
7.4 |
19 |
Fold:C2 domain-like
Superfamily:C2 domain (Calcium/lipid-binding domain, CaLB)
Family:PLC-like (P variant) |