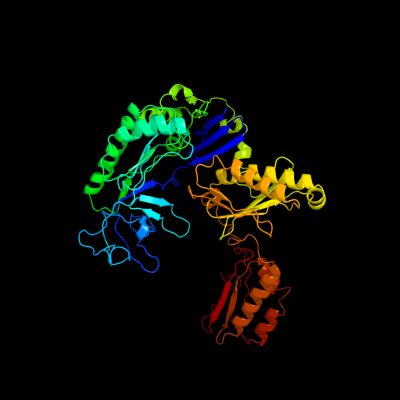
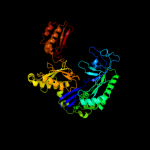
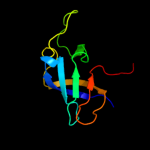
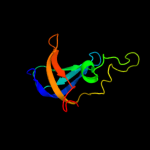
1 c2c4rL_
100.0
100
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
2 d1smxa_
99.9
100
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like3 d2z0sa1
98.3
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like4 c2cqoA_
98.2
16
PDB header: ribosomeChain: A: PDB Molecule: nucleolar protein of 40 kda;PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
5 d2ba0a1
98.2
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like6 c1yz6A_
98.1
24
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
7 d1q46a2
98.1
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like8 c2z0sA_
98.1
14
PDB header: rna binding proteinChain: A: PDB Molecule: probable exosome complex rna-binding protein 1;PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
9 c2k4kA_
98.1
23
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
10 d3bzka4
98.1
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like11 c2je6I_
98.0
19
PDB header: hydrolaseChain: I: PDB Molecule: exosome complex rna-binding protein 1;PDBTitle: structure of a 9-subunit archaeal exosome
12 c2khjA_
98.0
25
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
13 d2je6i1
98.0
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like14 c2khiA_
98.0
14
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
15 c2ba0A_
97.9
16
PDB header: rna binding proteinChain: A: PDB Molecule: archeal exosome rna binding protein rrp4;PDBTitle: archaeal exosome core
16 d2nn6h1
97.9
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like17 c1q8kA_
97.9
19
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
18 c2eqsA_
97.9
38
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dhx8;PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
19 d1wi5a_
97.9
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like20 d1sroa_
97.9
32
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like21 d1go3e1
not modelled
97.8
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like22 c1q46A_
not modelled
97.8
19
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
23 d1hh2p1
not modelled
97.7
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like24 c3go5A_
not modelled
97.6
12
PDB header: gene regulationChain: A: PDB Molecule: multidomain protein with s1 rna-binding domains;PDBTitle: crystal structure of a multidomain protein with nucleic acid binding2 domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 a3 resolution
25 c2oceA_
not modelled
97.6
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
26 c1hh2P_
not modelled
97.6
26
PDB header: transcription regulationChain: P: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima
27 c2ba1B_
not modelled
97.5
16
PDB header: rna binding proteinChain: B: PDB Molecule: archaeal exosome rna binding protein csl4;PDBTitle: archaeal exosome core
28 c2ahoB_
not modelled
97.4
27
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: structure of the archaeal initiation factor eif2 alpha-2 gamma heterodimer from sulfolobus solfataricus complexed3 with gdpnp
29 d2ahob2
not modelled
97.4
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like30 c1l2fA_
not modelled
97.4
27
PDB header: transcriptionChain: A: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
31 d2nn6i1
not modelled
97.4
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like32 d1kl9a2
not modelled
97.3
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like33 c2k52A_
not modelled
97.3
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
34 c1go3E_
not modelled
97.1
20
PDB header: transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
35 c3psiA_
not modelled
97.1
13
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
36 c2nn6I_
not modelled
97.0
19
PDB header: hydrolase/transferaseChain: I: PDB Molecule: 3'-5' exoribonuclease csl4 homolog;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
37 d1y14b1
not modelled
97.0
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like38 c2c35F_
not modelled
96.5
15
PDB header: polymeraseChain: F: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
39 c3h0gS_
not modelled
96.5
12
PDB header: transcriptionChain: S: PDB Molecule: dna-directed rna polymerase ii subunit rpb7;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
40 c1kl9A_
not modelled
96.4
21
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2 subunit 1;PDBTitle: crystal structure of the n-terminal segment of human eukaryotic2 initiation factor 2alpha
41 d2nn6g1
not modelled
96.3
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like42 c2b8kG_
not modelled
96.2
14
PDB header: transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: 12-subunit rna polymerase ii
43 d2c35b1
not modelled
95.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like44 c2pmzE_
not modelled
95.4
16
PDB header: translation, transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
45 c2bh8B_
not modelled
95.4
20
PDB header: transcriptionChain: B: PDB Molecule: 1b11;PDBTitle: combinatorial protein 1b11
46 c1nt9G_
not modelled
95.3
14
PDB header: transcription, transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kd polypeptide;PDBTitle: complete 12-subunit rna polymerase ii
47 c3aqqD_
not modelled
95.1
25
PDB header: dna binding proteinChain: D: PDB Molecule: calcium-regulated heat stable protein 1;PDBTitle: crystal structure of human crhsp-24
48 c2nn6H_
not modelled
94.7
15
PDB header: hydrolase/transferaseChain: H: PDB Molecule: exosome complex exonuclease rrp4;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
49 c2ckzB_
not modelled
94.6
13
PDB header: transferaseChain: B: PDB Molecule: dna-directed rna polymerase iii 25 kdPDBTitle: x-ray structure of rna polymerase iii subcomplex c17-c25.
50 d2asba1
not modelled
94.4
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like51 c2nn6G_
not modelled
94.2
19
PDB header: hydrolase/transferaseChain: G: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
52 c1k0rB_
not modelled
94.0
20
PDB header: transcriptionChain: B: PDB Molecule: nusa;PDBTitle: crystal structure of mycobacterium tuberculosis nusa
53 c2asbA_
not modelled
94.0
23
PDB header: transcription/rnaChain: A: PDB Molecule: transcription elongation protein nusa;PDBTitle: structure of a mycobacterium tuberculosis nusa-rna complex
54 d2ja9a1
not modelled
92.9
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like55 d1c9oa_
not modelled
92.5
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like56 d1h95a_
not modelled
91.3
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like57 c2ljpA_
not modelled
90.8
10
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease p protein component;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for e.coli2 ribonuclease p protein
58 d1mjca_
not modelled
90.5
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like59 d2es2a1
not modelled
90.5
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like60 c2kcmA_
not modelled
88.7
19
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: cold shock domain family protein;PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
61 d1g6pa_
not modelled
88.3
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like62 d1d6ta_
not modelled
87.7
21
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein63 d1nz0a_
not modelled
87.6
21
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein64 c2eqjA_
not modelled
86.9
19
PDB header: transcriptionChain: A: PDB Molecule: metal-response element-binding transcriptionPDBTitle: solution structure of the tudor domain of metal-response2 element-binding transcription factor 2
65 c2ja9A_
not modelled
86.2
14
PDB header: rna-binding proteinChain: A: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the n-terminal deletion of yeast exosome2 component rrp40
66 d1k3ra1
not modelled
85.8
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Hypothetical protein MTH1 (MT0001), insert domain67 c2hnkC_
not modelled
85.6
20
PDB header: transferaseChain: C: PDB Molecule: sam-dependent o-methyltransferase;PDBTitle: crystal structure of sam-dependent o-methyltransferase from2 pathogenic bacterium leptospira interrogans
68 c2wp8J_
not modelled
85.3
11
PDB header: hydrolaseChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: yeast rrp44 nuclease
69 c2e5pA_
not modelled
84.0
20
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 1 (phf1 protein)
70 c3d0fA_
not modelled
83.6
10
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
71 c2k5nA_
not modelled
83.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cold-shock protein;PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
72 c2pe4A_
not modelled
83.4
20
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronidase-1;PDBTitle: structure of human hyaluronidase 1, a hyaluronan hydrolyzing enzyme2 involved in tumor growth and angiogenesis
73 d1a6fa_
not modelled
82.7
16
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein74 c3trzE_
not modelled
81.5
25
PDB header: rna binding protein/rnaChain: E: PDB Molecule: protein lin-28 homolog a;PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
75 c3pihA_
not modelled
81.5
20
PDB header: hydrolase/dnaChain: A: PDB Molecule: uvrabc system protein a;PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
76 c3pnwX_
not modelled
81.0
23
PDB header: protein binding/immune systemChain: X: PDB Molecule: tudor domain-containing protein 3;PDBTitle: crystal structure of the tudor domain of human tdrd3 in complex with2 an anti-tdrd3 fab
77 c2bx9J_
not modelled
79.8
36
PDB header: transcription regulationChain: J: PDB Molecule: tryptophan rna-binding attenuator protein-inhibitoryPDBTitle: crystal structure of b.subtilis anti-trap protein, an2 antagonist of trap-rna interactions
78 c3a0jB_
not modelled
78.1
17
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
79 c2ytvA_
not modelled
77.2
20
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fifth cold-shock domain of the human2 kiaa0885 protein (unr protein)
80 c2e5qA_
not modelled
76.0
24
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 19;PDBTitle: solution structure of the tudor domain of phd finger2 protein 19, isoform b [homo sapiens]
81 c3ld0Q_
not modelled
75.1
30
PDB header: gene regulationChain: Q: PDB Molecule: inhibitor of trap, regulated by t-box (trp) sequence rtpa;PDBTitle: crystal structure of b.licheniformis anti-trap protein, an antagonist2 of trap-rna interactions
82 c2d9tA_
not modelled
73.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tudor domain-containing protein 3;PDBTitle: solution structure of the tudor domain of tudor domain2 containing protein 3 from mouse
83 c2atmA_
not modelled
73.6
23
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: crystal structure of the recombinant allergen ves v 2
84 d1fcqa_
not modelled
72.4
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Bee venom hyaluronidase85 c1g5vA_
not modelled
70.2
19
PDB header: translationChain: A: PDB Molecule: survival motor neuron protein 1;PDBTitle: solution structure of the tudor domain of the human smn2 protein
86 d1mhna_
not modelled
70.0
19
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain87 c1fcuA_
not modelled
69.6
26
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: crystal structure (trigonal) of bee venom hyaluronidase
88 c3cbgA_
not modelled
68.7
28
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase;PDBTitle: functional and structural characterization of a2 cationdependent o-methyltransferase from the3 cyanobacterium synechocystis sp. strain pcc 6803
89 d1nlta3
not modelled
67.9
28
Fold: DnaJ/Hsp40 cysteine-rich domainSuperfamily: DnaJ/Hsp40 cysteine-rich domainFamily: DnaJ/Hsp40 cysteine-rich domain90 c3camB_
not modelled
67.4
21
PDB header: gene regulationChain: B: PDB Molecule: cold-shock domain family protein;PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
91 c3ayhB_
not modelled
66.8
15
PDB header: transcriptionChain: B: PDB Molecule: dna-directed rna polymerase iii subunit rpc8;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
92 c3tr6A_
not modelled
65.4
30
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase;PDBTitle: structure of a o-methyltransferase from coxiella burnetii
93 d3d3ra1
not modelled
63.8
20
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like94 d1bi9a_
not modelled
63.2
19
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like95 d2ix0a2
not modelled
62.6
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like96 c3nthA_
not modelled
62.6
25
PDB header: transcriptionChain: A: PDB Molecule: maternal protein tudor;PDBTitle: crystal structure of tudor and aubergine [r13(me2s)] complex
97 c2hl7A_
not modelled
61.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
98 d1wfqa_
not modelled
58.1
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like99 c1nltA_
not modelled
57.8
21
PDB header: protein transportChain: A: PDB Molecule: mitochondrial protein import protein mas5;PDBTitle: the crystal structure of hsp40 ydj1
100 c1x65A_
not modelled
56.9
15
PDB header: rna binding proteinChain: A: PDB Molecule: unr protein;PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
101 c1tt9B_
not modelled
56.5
38
PDB header: transferase, lyaseChain: B: PDB Molecule: formimidoyltransferase-cyclodeaminasePDBTitle: structure of the bifunctional and golgi associated2 formiminotransferase cyclodeaminase octamer
102 c2hqxB_
not modelled
56.2
21
PDB header: transcriptionChain: B: PDB Molecule: p100 co-activator tudor domain;PDBTitle: crystal structure of human p100 tudor domain conserved2 region
103 d2hqxa1
not modelled
56.2
21
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain104 c3d3rA_
not modelled
55.4
20
PDB header: chaperoneChain: A: PDB Molecule: hydrogenase assembly chaperone hypc/hupf;PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
105 c2ix1A_
not modelled
55.4
16
PDB header: hydrolaseChain: A: PDB Molecule: exoribonuclease 2;PDBTitle: rnase ii d209n mutant
106 d1h9ma1
not modelled
54.5
19
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain107 d1vlua_
not modelled
54.4
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like108 d1a62a2
not modelled
53.4
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like109 d1h9ra1
not modelled
52.8
26
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain110 c2cttA_
not modelled
52.7
31
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of zinc finger domain from human dnaj2 subfamily a menber 3
111 c3bdlA_
not modelled
52.6
21
PDB header: hydrolaseChain: A: PDB Molecule: staphylococcal nuclease domain-containingPDBTitle: crystal structure of a truncated human tudor-sn
112 d2d9ta1
not modelled
52.1
26
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain113 d1exka_
not modelled
50.9
38
Fold: DnaJ/Hsp40 cysteine-rich domainSuperfamily: DnaJ/Hsp40 cysteine-rich domainFamily: DnaJ/Hsp40 cysteine-rich domain114 d1p6oa_
not modelled
50.1
14
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like115 c3lz8A_
not modelled
49.8
20
PDB header: chaperoneChain: A: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone dnaj from klebsiella pneumoniae2 subsp. pneumoniae mgh 78578 at 2.9 a resolution.
116 c4a4fA_
not modelled
49.5
26
PDB header: rna binding proteinChain: A: PDB Molecule: survival of motor neuron-related-splicing factor 30;PDBTitle: solution structure of spf30 tudor domain in complex with2 symmetrically dimethylated arginine
117 c2ytxA_
not modelled
49.3
15
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the second cold-shock domain of the human2 kiaa0885 protein (unr protein)
118 d1guta_
not modelled
49.2
17
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP119 c2eqkA_
not modelled
49.2
25
PDB header: transcriptionChain: A: PDB Molecule: tudor domain-containing protein 4;PDBTitle: solution structure of the tudor domain of tudor domain-2 containing protein 4
120 d2diqa1
not modelled
46.8
25
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain