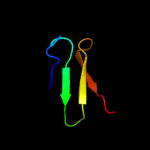
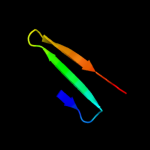
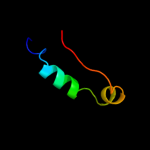
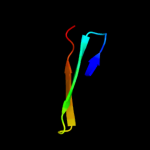
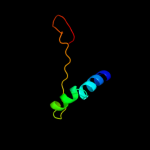
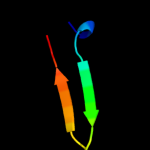
1 d1ew4a_
100.0
100
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like2 d1ekga_
100.0
25
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like3 d2ga5a1
100.0
27
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like4 d2fqla1
100.0
28
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like5 d1xkpb1
61.4
11
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone6 c1q40C_
32.2
43
PDB header: translationChain: C: PDB Molecule: mrna transport regulator mtr2;PDBTitle: crystal structure of the c. albicans mtr2-mex67 m domain complex
7 d1mbya_
29.1
13
Fold: Polo-box domainSuperfamily: Polo-box domainFamily: Swapped Polo-box domain8 d2ebsa1
23.9
24
Fold: 7-bladed beta-propellerSuperfamily: Oligoxyloglucan reducing end-specific cellobiohydrolaseFamily: Oligoxyloglucan reducing end-specific cellobiohydrolase9 d2ogqa2
22.1
26
Fold: Polo-box domainSuperfamily: Polo-box domainFamily: Polo-box duplicated region10 d3e11a1
21.0
15
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: TTHA0227-like11 d1q42a_
20.4
43
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: NTF2-like12 c1umwB_
19.5
22
PDB header: kinaseChain: B: PDB Molecule: serine/threonine-protein kinase plk;PDBTitle: structure of a human plk1 polo-box domain/phosphopeptide2 complex
13 c2bsgA_
18.8
20
PDB header: viral proteinChain: A: PDB Molecule: fibritin;PDBTitle: the modeled structure of fibritin (gpwac) of bacteriophage2 t4 based on cryo-em reconstruction of the extended tail of3 bacteriophage t4
14 c2v5iA_
18.1
24
PDB header: viral proteinChain: A: PDB Molecule: salmonella typhimurium db7155 bacteriophage det7PDBTitle: structure of the receptor-binding protein of bacteriophage2 det7: a podoviral tailspike in a myovirus
15 d2azeb1
14.4
23
Fold: E2F-DP heterodimerization regionSuperfamily: E2F-DP heterodimerization regionFamily: E2F dimerization segment16 c3aabA_
12.9
32
PDB header: chaperoneChain: A: PDB Molecule: putative uncharacterized protein st1653;PDBTitle: small heat shock protein hsp14.0 with the mutations of i120f and i122f2 in the form i crystal
17 d2h50a1
12.7
26
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP2018 c2v1lA_
12.4
19
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of the conserved hypothetical protein vc1805 from2 pathogenicity island vpi-2 of vibrio cholerae o1 biovar3 eltor str. n16961 shares structural homology with the4 human p32 protein
19 d1gmea_
11.6
26
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP2020 c3nrlB_
11.6
50
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rumgna_01417;PDBTitle: crystal structure of protein rumgna_01417 from ruminococcus gnavus,2 northeast structural genomics consortium target ugr76
21 c2qlvB_
not modelled
11.3
16
PDB header: transferase/protein bindingChain: B: PDB Molecule: protein sip2;PDBTitle: crystal structure of the heterotrimer core of the s.2 cerevisiae ampk homolog snf1
22 c3nmeA_
not modelled
11.3
16
PDB header: hydrolaseChain: A: PDB Molecule: sex4 glucan phosphatase;PDBTitle: structure of a plant phosphatase
23 c3btpA_
not modelled
11.1
33
PDB header: dna binding protein, chaperoneChain: A: PDB Molecule: single-strand dna-binding protein;PDBTitle: crystal structure of agrobacterium tumefaciens vire2 in complex with2 its chaperone vire1: a novel fold and implications for dna binding
24 d2qlvb1
not modelled
11.0
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like25 c3igmA_
not modelled
11.0
10
PDB header: transcription/dnaChain: A: PDB Molecule: pf14_0633 protein;PDBTitle: a 2.2a crystal structure of the ap2 domain of pf14_0633 from p.2 falciparum, bound as a domain-swapped dimer to its cognate dna
26 d1gmeb_
not modelled
10.6
26
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP2027 d2f15a1
not modelled
10.2
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like28 d2je8a2
not modelled
9.5
6
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain29 d1yuaa2
not modelled
8.9
18
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment30 d1z0mb1
not modelled
8.7
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like31 c2euzA_
not modelled
8.6
33
PDB header: cell cycle/dnaChain: A: PDB Molecule: ndt80 protein;PDBTitle: structure of a ndt80-dna complex (mse mutant mc5t)
32 d1mnna_
not modelled
8.6
33
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: DNA-binding domain from NDT8033 c3glaA_
not modelled
8.3
26
PDB header: chaperoneChain: A: PDB Molecule: low molecular weight heat shock protein;PDBTitle: crystal structure of the hspa from xanthomonas axonopodis
34 c1yuaA_
not modelled
7.8
18
PDB header: dna binding proteinChain: A: PDB Molecule: topoisomerase i;PDBTitle: c-terminal domain of escherichia coli topoisomerase i
35 d1z0na1
not modelled
7.6
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like36 c2yz0A_
not modelled
7.5
10
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase gcn2;PDBTitle: solution structure of rwd/gi domain of saccharomyces2 cerevisiae gcn2
37 c2vecA_
not modelled
7.4
19
PDB header: cytosolic proteinChain: A: PDB Molecule: pirin-like protein yhak;PDBTitle: the crystal structure of the protein yhak from escherichia2 coli
38 d1veha_
not modelled
6.8
18
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like39 d1xhja_
not modelled
6.7
8
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like40 c2pmzQ_
not modelled
6.1
28
PDB header: translation, transferaseChain: Q: PDB Molecule: dna-directed rna polymerase subunit a;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
41 d2p3ya1
not modelled
6.1
19
Fold: VPA0735-likeSuperfamily: VPA0735-likeFamily: VPA0735-like42 c2p3yA_
not modelled
6.1
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein vpa0735;PDBTitle: crystal structure of vpa0735 from vibrio parahaemolyticus. northeast2 structural genomics target vpr109
43 c3q9qB_
not modelled
6.0
16
PDB header: chaperoneChain: B: PDB Molecule: heat shock protein beta-1;PDBTitle: hspb1 fragment second crystal form
44 d1shsa_
not modelled
6.0
16
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP2045 c1shsD_
not modelled
6.0
16
PDB header: heat shock proteinChain: D: PDB Molecule: small heat shock protein;PDBTitle: small heat shock protein from methanococcus jannaschii
46 c3n23E_
not modelled
5.6
17
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
47 c2wj5A_
not modelled
5.5
21
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein beta-6;PDBTitle: rat alpha crystallin domain
48 c2k38A_
not modelled
5.5
40
PDB header: antimicrobial proteinChain: A: PDB Molecule: cupiennin-1a;PDBTitle: cupiennin 1a, nmr, minimized average structure
49 d1j1la_
not modelled
5.4
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Pirin-like50 c3k6qB_
not modelled
5.4
7
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
51 d1ro0a_
not modelled
5.3
36
Fold: Prim-pol domainSuperfamily: Prim-pol domainFamily: Bifunctional DNA primase/polymerase N-terminal domain52 c2x6vB_
not modelled
5.2
7
PDB header: transcription/dnaChain: B: PDB Molecule: t-box transcription factor tbx5;PDBTitle: crystal structure of human tbx5 in the dna-bound and dna-2 free form
53 d1p5ta_
not modelled
5.1
21
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Phosphotyrosine-binding domain (PTB)