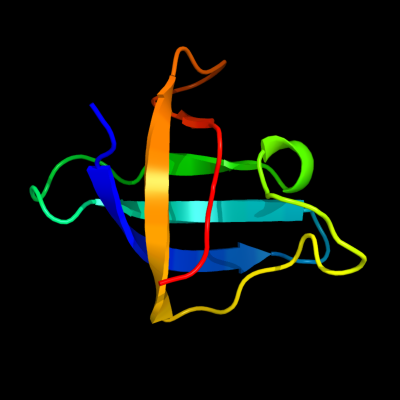
1 d1mjca_
99.9
69
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like2 d1h95a_
99.9
46
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like3 d2es2a1
99.9
65
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like4 d1c9oa_
99.9
64
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like5 c3camB_
99.9
64
PDB header: gene regulationChain: B: PDB Molecule: cold-shock domain family protein;PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
6 c3a0jB_
99.9
55
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
7 d1g6pa_
99.9
62
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like8 c2k5nA_
99.9
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cold-shock protein;PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
9 c2kcmA_
99.9
32
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: cold shock domain family protein;PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
10 c3aqqD_
99.8
36
PDB header: dna binding proteinChain: D: PDB Molecule: calcium-regulated heat stable protein 1;PDBTitle: crystal structure of human crhsp-24
11 c3trzE_
99.8
42
PDB header: rna binding protein/rnaChain: E: PDB Molecule: protein lin-28 homolog a;PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
12 c2ytyA_
99.7
28
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fourth cold-shock domain of the human2 kiaa0885 protein (unr protein)
13 d1wfqa_
99.7
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like14 c2ytxA_
99.7
32
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the second cold-shock domain of the human2 kiaa0885 protein (unr protein)
15 c1x65A_
99.6
29
PDB header: rna binding proteinChain: A: PDB Molecule: unr protein;PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
16 c2bh8B_
99.6
62
PDB header: transcriptionChain: B: PDB Molecule: 1b11;PDBTitle: combinatorial protein 1b11
17 c2ytvA_
99.5
32
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fifth cold-shock domain of the human2 kiaa0885 protein (unr protein)
18 d2ix0a2
97.8
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like19 c2ix1A_
97.0
25
PDB header: hydrolaseChain: A: PDB Molecule: exoribonuclease 2;PDBTitle: rnase ii d209n mutant
20 d1a62a2
96.8
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like21 c2a8vA_
not modelled
96.8
26
PDB header: protein/rnaChain: A: PDB Molecule: rna binding domain of rho transcriptionPDBTitle: rho transcription termination factor/rna complex
22 d1smxa_
not modelled
95.7
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like23 d2ix0a1
not modelled
94.4
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like24 c2vnuD_
not modelled
92.5
27
PDB header: hydrolase/rnaChain: D: PDB Molecule: exosome complex exonuclease rrp44;PDBTitle: crystal structure of sc rrp44
25 d1kl9a2
not modelled
89.9
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like26 c2k52A_
not modelled
89.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
27 d1y14b1
not modelled
89.4
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like28 c1hh2P_
not modelled
87.9
17
PDB header: transcription regulationChain: P: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima
29 c1l2fA_
not modelled
87.9
17
PDB header: transcriptionChain: A: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
30 c1xpuB_
not modelled
87.7
26
PDB header: transcription/rnaChain: B: PDB Molecule: rho transcription termination factor;PDBTitle: structural mechanism of inhibition of the rho transcription2 termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-3 dihydrobicyclomycin (fpdb)
31 c3go5A_
not modelled
86.0
13
PDB header: gene regulationChain: A: PDB Molecule: multidomain protein with s1 rna-binding domains;PDBTitle: crystal structure of a multidomain protein with nucleic acid binding2 domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 a3 resolution
32 d1u0la1
not modelled
85.9
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like33 d2c35b1
not modelled
84.7
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like34 c2cqoA_
not modelled
83.5
18
PDB header: ribosomeChain: A: PDB Molecule: nucleolar protein of 40 kda;PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
35 d1hh2p1
not modelled
82.7
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like36 c2l55A_
not modelled
79.0
25
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
37 c1u0lB_
not modelled
76.7
18
PDB header: hydrolaseChain: B: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of yjeq from thermotoga maritima
38 c1zeqX_
not modelled
76.2
12
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
39 c2c4rL_
not modelled
75.2
24
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
40 d2z0sa1
not modelled
74.2
9
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like41 c2wp8J_
not modelled
73.8
21
PDB header: hydrolaseChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: yeast rrp44 nuclease
42 c2khiA_
not modelled
73.7
23
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
43 d2nn6h1
not modelled
72.0
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like44 d1wi5a_
not modelled
70.1
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like45 c2oceA_
not modelled
69.6
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
46 c3l0oB_
not modelled
66.2
29
PDB header: hydrolaseChain: B: PDB Molecule: transcription termination factor rho;PDBTitle: structure of rna-free rho transcription termination factor from2 thermotoga maritima
47 d2ba0a1
not modelled
61.0
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like48 c3h0gS_
not modelled
60.5
25
PDB header: transcriptionChain: S: PDB Molecule: dna-directed rna polymerase ii subunit rpb7;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
49 d1go3e1
not modelled
57.8
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like50 c2khjA_
not modelled
56.9
16
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
51 d1q46a2
not modelled
56.3
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like52 d2asba1
not modelled
54.0
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like53 c2k4kA_
not modelled
51.9
21
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
54 d3bzka4
not modelled
49.5
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like55 c2c35F_
not modelled
49.1
18
PDB header: polymeraseChain: F: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
56 d2je6i1
not modelled
48.8
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like57 c2eqsA_
not modelled
43.8
20
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dhx8;PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
58 c1nt9G_
not modelled
43.7
22
PDB header: transcription, transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kd polypeptide;PDBTitle: complete 12-subunit rna polymerase ii
59 c2z0sA_
not modelled
43.6
6
PDB header: rna binding proteinChain: A: PDB Molecule: probable exosome complex rna-binding protein 1;PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
60 d2f3ga_
not modelled
43.0
33
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like61 c2b8kG_
not modelled
41.8
24
PDB header: transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: 12-subunit rna polymerase ii
62 d1glaf_
not modelled
38.2
33
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like63 d1j6qa_
not modelled
37.8
20
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE64 c1j6qA_
not modelled
37.8
20
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c maturation protein e;PDBTitle: solution structure and characterization of the heme2 chaperone ccme
65 c1q46A_
not modelled
37.2
11
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
66 c2xnqA_
not modelled
37.1
32
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein 3;PDBTitle: structural insights into cis element recognition of non-2 polyadenylated rnas by the nab3-rrm
67 c2pmzE_
not modelled
36.7
24
PDB header: translation, transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
68 d1h9ma2
not modelled
34.2
16
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain69 c2dhxA_
not modelled
32.9
21
PDB header: rna binding proteinChain: A: PDB Molecule: poly (adp-ribose) polymerase family, member 10PDBTitle: solution structure of the rrm domain in the human poly (adp-2 ribose) polymerase family, member 10 variant
70 d1gpra_
not modelled
32.3
27
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like71 c2gu1A_
not modelled
29.3
20
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
72 d2gpra_
not modelled
29.2
20
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like73 c3ggeA_
not modelled
26.9
16
PDB header: protein bindingChain: A: PDB Molecule: pdz domain-containing protein gipc2;PDBTitle: crystal structure of the pdz domain of pdz domain-containing protein2 gipc2
74 c2asbA_
not modelled
26.5
11
PDB header: transcription/rnaChain: A: PDB Molecule: transcription elongation protein nusa;PDBTitle: structure of a mycobacterium tuberculosis nusa-rna complex
75 c2qt7B_
not modelled
26.5
30
PDB header: hydrolaseChain: B: PDB Molecule: receptor-type tyrosine-protein phosphatase-likePDBTitle: crystallographic structure of the mature ectodomain of the2 human receptor-type protein-tyrosine phosphatase ia-2 at3 1.30 angstroms
76 d2vnud3
not modelled
26.0
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like77 c3psiA_
not modelled
25.5
13
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
78 c1q8kA_
not modelled
25.3
14
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
79 c2kjdA_
not modelled
24.8
14
PDB header: signaling proteinChain: A: PDB Molecule: sodium/hydrogen exchange regulatory cofactor nhe-PDBTitle: solution structure of extended pdz2 domain from nherf1 (150-2 270)
80 c2ahoB_
not modelled
24.7
20
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: structure of the archaeal initiation factor eif2 alpha-2 gamma heterodimer from sulfolobus solfataricus complexed3 with gdpnp
81 c1yz6A_
not modelled
24.5
24
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
82 d1ueqa_
not modelled
23.8
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain83 c3soeA_
not modelled
23.7
19
PDB header: signaling proteinChain: A: PDB Molecule: membrane-associated guanylate kinase, ww and pdz domain-PDBTitle: crystal structure of the 3rd pdz domain of the human membrane-2 associated guanylate kinase, ww and pdz domain-containing protein 33 (magi3)
84 d1h9ma1
not modelled
23.4
18
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain85 d1t9ha1
not modelled
22.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like86 c2je6I_
not modelled
22.3
18
PDB header: hydrolaseChain: I: PDB Molecule: exosome complex rna-binding protein 1;PDBTitle: structure of a 9-subunit archaeal exosome
87 d1guta_
not modelled
21.3
16
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP88 c1go3E_
not modelled
21.0
31
PDB header: transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
89 d1be9a_
not modelled
20.9
24
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain90 c1h9mB_
not modelled
20.8
16
PDB header: binding proteinChain: B: PDB Molecule: molybdenum-binding-protein;PDBTitle: two crystal structures of the cytoplasmic molybdate-binding2 protein modg suggest a novel cooperative binding mechanism3 and provide insights into ligand-binding specificity.4 peg-grown form with molybdate bound
91 d2ahob2
not modelled
20.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like92 d1fr3a_
not modelled
20.4
14
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP93 c2dc2A_
not modelled
20.4
16
PDB header: structural proteinChain: A: PDB Molecule: golgi associated pdz and coiled-coil motifPDBTitle: solution structure of pdz domain
94 d1sroa_
not modelled
19.8
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like95 c3bpuA_
not modelled
19.2
21
PDB header: transferaseChain: A: PDB Molecule: membrane-associated guanylate kinase, ww and pdz domain-PDBTitle: crystal structure of the 3rd pdz domain of human membrane associated2 guanylate kinase, c677s and c709s double mutant
96 d1wf1a_
not modelled
18.6
26
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD97 d1ujva_
not modelled
18.4
21
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain98 c2kviA_
not modelled
18.2
32
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein 3;PDBTitle: structure of nab3 rrm
99 d2vpaa1
not modelled
18.1
11
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like